- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Discussions
Solve problems, and share tips and tricks with other JMP users.- JMP User Community
- :
- Discussions
- :
- Re: Definitive Screening Design Workflow
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Definitive Screening Design Workflow
Hello,
I ran a Definitive screening design and generated a table by selecting "Add block with center runs to estimate quadratic effects. I collected the response data and added it to the table. But when I try to run "Fit Definitive Screening", I'm met with this error:
The Fit Definitive Screening platform only runs when I hide/exclude runs from block 2. Am I doing something wrong? How do I run a model with all of the runs included? Do I have do some kind of augmented design to include the second block?
Accepted Solutions
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Definitive Screening Design Workflow
Hi @sanch1,
Sorting the datatable don't change the design structure and analysis. However, removing an experiment from the table will destroy the design foldover structure (there will be one experimental run without its "mirror image" counterpart, which prevent from using the Fit DSD analysis platform). You will face the same problem and error message with response missing value(s), or by excluding row(s) in the table.
I have the same error message than you when removing any run from the design table and trying to launch the Fit DSD platform :
I would recommend using other modeling platforms (as mentioned in my response before) to do the analysis.
Hope this answer your question,
"It is not unusual for a well-designed experiment to analyze itself" (Box, Hunter and Hunter)
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Definitive Screening Design Workflow
Hi @sanch1,
I just tried to reproduce the same error by creating the same design type with 8 continuous factors, 2 blocks, 8 extra runs :
When using a random formula to model a response with this design, I have no error and can continue the analysis :
I have the same design as you (you can also find it attached) :
Normally you would get this error message when one or several experimental runs are not part of the design and destroys the foldover structure, see https://community.jmp.com/t5/Discussions/How-can-I-add-extra-runs-into-the-designed-runs-from-Defini...
There are many other ways to proceed with the analysis using the Fit Model platform if necessary, using Stepwise models, Generalized regression models, etc...
But in your case, row 18 is perfectly fine and is the "mirror image" of row 17 in the same block 1, so the foldover structure is present and respected.
Did you change any factors values in this row (or in others) after generating the design ?
Did you try launching the "Fit DSD" platform from the menu (menu DOE, Definitive Screening, Fit Definitive Screening) ?
Can you share an anonymized version of your dataset and design ?
Hope this first discussion starter might help you,
"It is not unusual for a well-designed experiment to analyze itself" (Box, Hunter and Hunter)
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Definitive Screening Design Workflow
Hi Victor,
I did make some changes to the table:
1. I sorted the data table ascending by block for organization
2. The experiment in run 19 I had to remove due to it failing before I could collect results.
would either of these have threw off the DSD design?
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Definitive Screening Design Workflow
Hi @sanch1,
Sorting the datatable don't change the design structure and analysis. However, removing an experiment from the table will destroy the design foldover structure (there will be one experimental run without its "mirror image" counterpart, which prevent from using the Fit DSD analysis platform). You will face the same problem and error message with response missing value(s), or by excluding row(s) in the table.
I have the same error message than you when removing any run from the design table and trying to launch the Fit DSD platform :
I would recommend using other modeling platforms (as mentioned in my response before) to do the analysis.
Hope this answer your question,
"It is not unusual for a well-designed experiment to analyze itself" (Box, Hunter and Hunter)
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Definitive Screening Design Workflow
I went ahead using a method with the All possible models approach through Stepwise. Thank you so much for your help!
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Definitive Screening Design Workflow
Pardon my comments (and ignore them if you prefer), but stepwise is not how you should analyze DOE. Stepwise is an additive model building approach useful when you don't have a model in mind (e.g., data mining). When you run experiments you start with a model in mind. This is a function of the factors and levels (and strategies for handling noise). From the model you started with, you remove insignificant terms (a subtractive approach).
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Definitive Screening Design Workflow
Interesting approach! However, given this particular approach where I have one data point (as I wasn't able to complete that run), what would you recommend instead to get anything useful out of the data I still have?
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Definitive Screening Design Workflow
Sorry, I haven't been following the whole thread and I saw Victor was giving advice so I didn't comment. You ran an experiment with a set number of factors and levels. Apparently you also decided to run blocks and center points. Regardless, you should have a model assigning your DF's. And you should absolutely analyze the data with that model.
See my advice from this thread regarding missing data:
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Definitive Screening Design Workflow
Hi @statman,
Thanks a lot for your input and comments, always instructive and thoughtful to guide new and experienced users.
I tend to agree with you about the Stepwise approach on the "theoritical" aspect: for non-(super)saturated designs, the assumed model should be the one you're starting with, before considering refining it based on statistical criteria and practical evaluation/validation.
With Definitive Screening Designs, the situation tends to be a little different compared to traditional designs, since you won't be able to estimate all possible terms that could be estimated and enter in the model ; in the situation here with 8 factors and 1 block, that would mean to estimate 1 intercept, 8 main effects (+block), 28 interactions between 2 factors, and 8 quadratic effects. The design used here uses 26 runs, so it can't estimate a full RSM model with the 46 terms mentioned before, so no possible backwards/subtractive approach possible without strong assumptions/simplification.
Hence the need for a specific analysis strategy, which is under the "Fit DSD" platform. If possible, the "Fit DSD" analysis is the recommended analysis, as it is a more conservative analysis strategy than Stepwise approaches, assuming factor sparsity and effect heredity principles hold true, estimating and fitting main effects first, before considering interactions and quadratic effects with effect heredity principle and estimating them from the residuals of the main effects model.
When Fit DSD is not possible (because of missing values, excluded rows, added replicates, ... anything that could destroy the foldover structure and prevent fom using the recommended analysis approach for DSD), then you have to find something else in practice. Stepwise may be an option (as well as Generalized Regression models, with "Two Stage Forward Selection", "Pruned Forward Selection" or "Best Subset" estimation methods with Effect Heredity enforced, but only available in JMP Pro), even if its "brute-force" and greedy approach may not be optimal in the context of designed experiments.
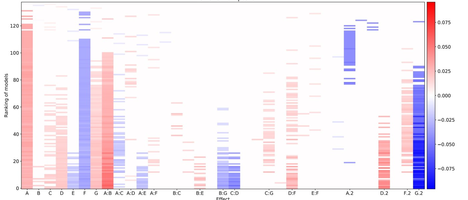
I particularly like the "All Models" option in the Stepwise platform (for limited number of factors and terms in the model), not to directly create in a brute-force approach the "best" model, but to guide the understanding and evaluation of several models, and choose the most likely active terms in the final model. This can be visualized through " Raster plots", introduced in the context of model selection for DoE by Peter Goos, proposed in the JMP Wish List : Raster plots or other visualization tools to help model evaluation and selection for DoEs
This visualization helps to identify the most likely active terms, and see where/how models agree or disagree. It can also help visualizing aliasing between effects. Example from a use case by Peter Goos :
@sanch1 At the end, "All models are wrong but some are useful", so it's always interesting to try and compare different modeling options, and even more when domain expertise can guide the process. Some methods are more conservative than others, but combining different modeling with domain expertise can help having a broader view about what matters the most. And from then, plan your next experiments to augment your DoE, confirm/refine/correct your model, and prepare some validation points to be able to assess your model's validity.
If you need more informations or are interested in diving deeper in the analysis of DSD topic, there are other ressources/posts that could help you :
- More infos about the Fit DSD : Information tools for analysis of Definitive Screening Designs
- Discussion on analytical strategies and differences between Fit DSD and Stepwise approaches : Fit Definitive Screening vs. Stepwise (min. AICC) for model selection
I hope this complementary answer may be helpful,
"It is not unusual for a well-designed experiment to analyze itself" (Box, Hunter and Hunter)
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Definitive Screening Design Workflow
Well we have to agree to disagree. I have attached the DSD you attached earlier in the thread with a saturated model (fun the Fit Model script). From there you subtract terms.
Just because you aren't assigning the DF's doesn't mean they cab't be (agreeably randomized replicates do not allow for assignment)
Recommended Articles
- © 2026 JMP Statistical Discovery LLC. All Rights Reserved.
- Terms of Use
- Privacy Statement
- Contact Us