- JMP will suspend normal business operations for our Winter Holiday beginning on Wednesday, Dec. 24, 2025, at 5:00 p.m. ET (2:00 p.m. ET for JMP Accounts Receivable).
Regular business hours will resume at 9:00 a.m. EST on Friday, Jan. 2, 2026. - We’re retiring the File Exchange at the end of this year. The JMP Marketplace is now your destination for add-ins and extensions.
- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Discussions
Solve problems, and share tips and tricks with other JMP users.- JMP User Community
- :
- Discussions
- :
- Re: RNAi screening
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
RNAi screening
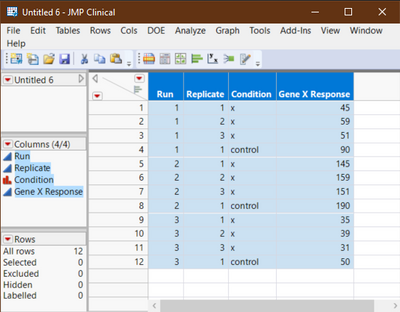
I have an RNAi dataset with 3 technical replicates that was done in triplicate. For example:
Run 1
gene X 45
gene X 59
gene X 51
Control 90
Run 2
gene X 145
gene X 159
gene X 151
Control 190
Run 3
gene X 35
gene X 39
gene X 31
Control 50
The real dataset has hundreds of genes and dozens of controls per experiment. Each run tends to vary in its average, but the experiments were the same. I think this is due to room humidity, but it doesn't matter in the end. My interest is in the genes, not the run variation. I basically want to see which genes vary from control, and a probability (p value?). What is the best approach to analyzing this data with JMP?
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: RNAi screening
Hi @thestrider
I would happy to guide you through this process.
I have some clarification/verification questions since my ultimate response will depend on these answers.
- So does your table of data look something like this but with many genes?
- Is there only one control per run but three technical measurements of the same condition within the same run (like in the table above)?
- Do you have something like condition x is one experimental condition, with three technical replicates for gene x, and you have many conditions for the experiment (condition x, y, z, etc.)?
- As many details as you can give the more accurate and appropriate my answer/solution will be. Even what the output of your data looks like helps.
Thanks,
Data Scientist, Life Sciences - Global Technical Enablement
JMP Statistical Discovery, LLC. - Denver, CO
Tel: +1-919-531-9927 ▪ Mobile: +1-303-378-7419 ▪ E-mail: chris.kirchberg@jmp.com
www.jmp.com
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: RNAi screening
Hi Chris!
You are correct, my dataset looks like almost exactly like your picture, with 400 genes (gene x, y, z, etc) and some more metadata that is just for validating. (ie 96 well microplate location, the plate name etc)
Each run has many controls, some are empty vector, (baseline, no effect) others are known genes that generate high or low responses. Every microplate had controls, and we also just tested a control only microplate in each of the three runs. About 15% of my dataset (~2000 of 15000 measurements) is controls.
- Do you have something like condition x is one experimental condition, with three technical replicates for gene x, and you have many conditions for the experiment (condition x, y, z, etc.)
I think this is a good way to describe it. Each gene in the list is a gene that was turned off. So I turned off 400 genes, one at a time. In each run I had three identical copies of each microplate, so there are 3 technical replicates of each gene. I should note here that randomly about 15% of data was censored out (bacteria didn't grow, contaminated, etc), so sometimes genes do not have 3 technical replicates. I then repeated this entire experiment the same way two more times. So in an ideal world I have 9 data points per condition. Censoring is random though so some genes have only 4-5 data points.
The empty vector control average varies across all three experiments. But the response of each gene should be similar. So a gene that lowers the value from baseline should always lower it, but the numbers themselves vary sometimes drastically in the three runs. So gene X might have a value of say 70 vs a 100 average baseline in one run, but a value of 120 vs a baseline of 300 in the next. I thought I might need to normalize, but I am worried that the data may be tailed and I am uncertain on the stats to do that. I am measuring fluorescent levels, and basically dim things don't get much dimmer but bright things can get very bright. So by tailed I mean in the example I just gave, a bright gene might be 350 vs 100 baseline, but in the next run it could be 1200 vs 300 baseline. In other words there is a big numerical range on the high end of the scale, but the low end there is a smaller range. I hope that makes sense.
I attached a little powerpoint, it shows what I am measuring, ( c. elegans nematodes) and a sampling of data.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: RNAi screening
Thanks @thestrider,
Yes, there are multiple things that need to be addressed here. I can't tell you what you should do statistically (which analysis you would need to do or how to address the between run variability due to external factors, like the shifting of control and treatment values) since we are advised to not give statistical advice on how to treat each of these issues, but I can point you to which platforms within JMP that could help address some of them
Within run statistical comparisons cannot be made since there is only one control per run and no way to assess variability for within run variation to make that comparison (at least not by traditional statistical methods that I am aware of). Between run variation can be estimated since there are three controls, but it will be biased because you can have up to 9 possible measurements for the condition of interest. This would be considered unbalanced and possibly give you unequal variance. Thankfully, JMP has a means to deal with unequal variance in the Fit Y by X platform (gene X response in the Y, Response section and condition in the X, Factor section). Then choose t-Test from the red triangle which will give you the means for each group and a p-value for testing if the condition is different from control, assuming unequal variance. The censored data is basically missing data (not measurable due to the reason given) and makes sense why you have added technical replicates to accommodate such issues for each run.
Also, as you have mentioned, values shift from one run to the other, including the control, almost like a match pair situation (or paired t-test like situation) or a batch effect. So the above alone is not enough to see if there is a statistical difference because of this additional bias. There could be many ways to address this. One may be to do multiple regression where run and condition are effects (kind of like batch and condition looking for batch effects and accommodating for them).
In this case, Fit Model is the platform to use and choosing Standard Least Squares as the personality will get you to the type of test/analysis that might be most appropriate in your case. I would first test to see if there is a run effect by putting run, condition and the interaction of run and condition as model effects and then Gene X response as the Y. You could do this for all genes (and that would be a lot to look at for hundreds of genes). The small example data you have, I have done this and there is definitely an effect due to run to run variability, so that means it must be accounted for in the statistical tests you perform.
I have created a JMP table and saved the Fit Y by X and the Fit Model analyses to the the data table so you can see how I performed each test. It is attached to this post.
It is best to either contact a statistician in house or see if one of our gracious community members are willing to advise on how best to treat your situation. Maybe also take the free Statistical Thinking for Industrial Problem Solving course might help learn more about statistics that could help in this situation. Or take a look at the Statistics Knowledge Portal for more statistical upskilling.
Sorry I am not able to give the statistical advice you might need, but I hope I at least pointed to where in JMP you could go to analyze your data and assess the experiment and test for significance of the conditions.
Best,
Data Scientist, Life Sciences - Global Technical Enablement
JMP Statistical Discovery, LLC. - Denver, CO
Tel: +1-919-531-9927 ▪ Mobile: +1-303-378-7419 ▪ E-mail: chris.kirchberg@jmp.com
www.jmp.com
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: RNAi screening
@Chris_Kirchberg wrote:
Within run statistical comparisons cannot be made since there is only one control per run and no way to assess variability for within run variation to make that comparison
Hi Chris, thanks so much for this that is incredibly helpful. I am a little confused by this part, sorry I was unclear, but I have ~210 empty vector control data points for each of my three runs. They are spread across every microplate I did (18 plates per run) but are identical otherwise. I also have a similar number of controls using known high and low responders.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: RNAi screening
Hi @thestrider,
Thanks for the clarification. I thought you only had one control measurement per gene per run. This makes more sense now.
I take it that the 400 genes are spread out among the 18 plates used in each run as well? Is each technical replicate for each gene placed on a different plate? So that means either 11 or 12 empty controls per plate and about 22 individual genes tested per plate. Is that correct?
The approach to analyzing this depends on the design of the experiment and placement of each technical replicate and the genes across the plates as well as how the empty (mock), low and high responders are to be used. How to assign the right variables to the right fields in JMP (such as Fit Model, Fit Y by Y, or even Response Screening) will depend on that. Same for how to structure the data table.
Since the controls are not gene specific, but a pool of empty/mock vector controls, that means the data table has to be structured such that the same pool of controls have to be reference for each gene. Also, there is going to be some normalization between plates and between runs as well.
Maybe someone on the community or at your organization can advise how to normalize this data and how to structure it so that one can use either Fit Model or Response Screening to use for your data. Those are the two platforms I would probably use to analyze the data in this case.
Sorry I could not guide you to the finer details of how you would approach the analysis.
Best,
Data Scientist, Life Sciences - Global Technical Enablement
JMP Statistical Discovery, LLC. - Denver, CO
Tel: +1-919-531-9927 ▪ Mobile: +1-303-378-7419 ▪ E-mail: chris.kirchberg@jmp.com
www.jmp.com
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: RNAi screening
@Chris_Kirchberg wrote:Hi @thestrider,
I take it that the 400 genes are spread out among the 18 plates used in each run as well? Is each technical replicate for each gene placed on a different plate? So that means either 11 or 12 empty controls per plate and about 22 individual genes tested per plate. Is that correct?
The approach to analyzing this depends on the design of the experiment and placement of each technical replicate and the genes across the plates as well as how the empty (mock), low and high responders are to be used. How to assign the right variables to the right fields in JMP (such as Fit Model, Fit Y by Y, or even Response Screening) will depend on that. Same for how to structure the data table.
You are close, each plate has 11-12 controls, and ~80 genes spread across 5 different microplates. A sixth microplate is just controls scattered across a plate. 3 copies of each plate were analyzed per run, for 18 plates total. Each technical replicate for each gene is thus on 3 plates. The empty vector controls are most important, they were to be used to determine variance in my experiments and establish a baseline response rate.
The way the experiment works- animals are fed a bacteria. In empty vector, that's it. But each gene represents a bacterial clone that turns off that specific gene. I can then measure the fluorescence of a reporter to determine how turning off that gene affects a pathway.
The high and low responders are there for two reasons- 1: To establish that the experiment is actually working and 2: to get a rough idea of the level of response we can expect from my assay. The high and low responders are the primary inhibitors and activators of the gene pathway I am studying, at least as far as my field currently knows. The other 400 genes are suspected of having an effect on this pathway in a prior experiment.
Since the controls are not gene specific, but a pool of empty/mock vector controls, that means the data table has to be structured such that the same pool of controls have to be reference for each gene. Also, there is going to be some normalization between plates and between runs as well.
Correct- the empty vector controls are a general reference for each gene. I think normalization between runs is very necessary. Between plates could be helpful as well, but the variance isn't as large there from what I see, at least using rough averages. If there is a way to quickly check for this in jmp I would love to know how to do it.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: RNAi screening
Just from an experimental design perspective, I'd like to get at that the original question was. It sounds like you are interested in determining how a gene responds relative to a control. But you have multiple types of controls, and I'm not clear what the goal of each control was. For example, I might subtract the true negative control average from the entire plate, to subtract the background, then I might normalize the genes on the plate to a house keeping gene to determine if the gene's signal was up or down regulated relative to something that is constitutively expressed. Or maybe normalize to a gene that is maximally expressed...
The additional problem here is that it sounds like each of the 18 plates has a wildly different dynamic range. And in addition to that noise, the dynamic range across the repeated experiments is also considerable.
One approach, and I'm sure there will be critics, is to preform background subtraction and normalization for each plate, Then either center and scale the response, or scale the response to 0-1. This forces the plate dynamic range to be identical for each plate.
Now that we have that problem tackled, assuming there is still something to analyze, we could look at the gene averages across the plates in each experimental replicate, and then take the average of the genes from the experimental replicates (yep double dipping on central limits theorem, and its legit.)
It sounds like you only have 3 or 4 technical replicates, so estimating sigma is a little sketchy with that sample size, but I might look at the %CV, and set some threshold for determining if the data for each gene is interpretable.
Granted all of this advise is based on having no idea how or what you did experimentally or even what the actual data even vaguely looks like. So this might be helpful, or completely useless, but hopefully the former.
-B
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: RNAi screening
First, thanks so much for all the replies, I am kind of floored by all the help I'm getting from this community, its amazing.
@Byron_JMP wrote:Just from an experimental design perspective, I'd like to get at that the original question was. It sounds like you are interested in determining how a gene responds relative to a control. But you have multiple types of controls, and I'm not clear what the goal of each control was. For example, I might subtract the true negative control average from the entire plate, to subtract the background, then I might normalize the genes on the plate to a house keeping gene to determine if the gene's signal was up or down regulated relative to something that is constitutively expressed. Or maybe normalize to a gene that is maximally expressed...
The goal was to discover genes that have an effect on a pathway using a fluorescent reporter. The empty vector is my baseline, it shows what happens when I don't do anything. So roughly analogous to using a housekeeping gene. The positive and negative are just there to see if my RNA interference is working (inducing the knockdown of a specific gene) , and to get a rough idea of the range of fluorescence in my assay. How would I do the normalization to empty vector using JMP? This is something I also thought would be helpful and I am glad to hear you think similarly.
The additional problem here is that it sounds like each of the 18 plates has a wildly different dynamic range. And in addition to that noise, the dynamic range across the repeated experiments is also considerable.
Note- the 18 plates in a run don't have wildly different dynamic range. It looks to be reasonable between plates, and I don't see much difference based on plate location either. The drastic dynamic range change is really seen when I replicated the experiment twice (I have 54 plates of data). The replicates were supposed to be identical but I think there was a humid day. My reporter is sensitive to oxygen...
One approach, and I'm sure there will be critics, is to preform background subtraction and normalization for each plate, Then either center and scale the response, or scale the response to 0-1. This forces the plate dynamic range to be identical for each plate.
Now that we have that problem tackled, assuming there is still something to analyze, we could look at the gene averages across the plates in each experimental replicate, and then take the average of the genes from the experimental replicates (yep double dipping on central limits theorem, and its legit.)
This sounds interesting, how would I do it in jmp?
It sounds like you only have 3 or 4 technical replicates, so estimating sigma is a little sketchy with that sample size, but I might look at the %CV, and set some threshold for determining if the data for each gene is interpretable.
I have up to 9 data points, 1 point from 3 microplates (technical replicates) each of three identical experiment days. (biological replicates) For this type of screen, in my field there are published papers with no replicates. I was hoping that by generating at least some replicates, I could catch weaker responders in my dataset. How can I set a threshold in jmp?
thanks, this is really really helpful!
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: RNAi screening
HI @thestrider ,
The tools in JMP you would need to use, probably, are Column Formulas (for creating normalized data to analyze) and Fit Model (standard least squares personality where you define your response and the model effects, random variables, interactions, etc.). How you will go about creating a formula for normalizing data and which model effects you will need for your experiment depend on you (your knowledge of statistics and your knowledge of the experimental design).
Here is a link to a search regarding analyzing 96 well plate data and designing experiments for 96 well plate formats within this community site. Take a look and it should provide some guidance on how to analyze your type of data given the format.
Also, there are some links to JMP's Documentation about creating Column Formulas and Fit Model using Standard Least Squares.
I hope that helps point you to where in JMP to perform these tasks.
Cheers,
Data Scientist, Life Sciences - Global Technical Enablement
JMP Statistical Discovery, LLC. - Denver, CO
Tel: +1-919-531-9927 ▪ Mobile: +1-303-378-7419 ▪ E-mail: chris.kirchberg@jmp.com
www.jmp.com
Recommended Articles
- © 2025 JMP Statistical Discovery LLC. All Rights Reserved.
- Terms of Use
- Privacy Statement
- Contact Us