- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Discussions
Solve problems, and share tips and tricks with other JMP users.- JMP User Community
- :
- Discussions
- :
- Re: Letters report vs differences report. Are they different?
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Letters report vs differences report. Are they different?
Hi,
I'm running a mixed model with JMP, and I just saw that letters from a tukey table don't match with their p-values from the differences report. I'm attaching a screen shoot about it. The JMP difference report matches with the Tukey-Kramer report from SAS too, but the Tukey table from JMP doesn't match with anything. Please I need help to understand what it is going on here and to know if I should use one of these separation tests at all.
Thanks,
Cristian
Accepted Solutions
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Letters report vs differences report. Are they different?
This discrepancy was reported before so it is a known bug. It was fixed in the JMP 13.2 maintenance release, so an update would fix it for you.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Letters report vs differences report. Are they different?
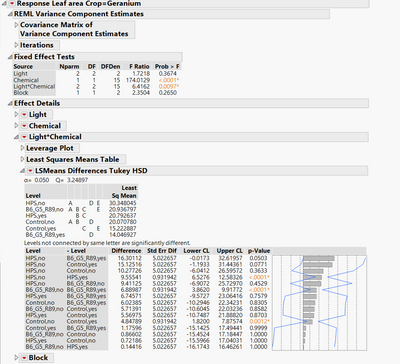
I found two discrepancies between the connecting letters report (LSMeans Differences Tukey HSD) and the p-values in the pair-wise report:
- HPS,yes (B,C) versus B6_G5_R89,yes (D) do not connect but p-value is 0.7579.
- Control,yes (C,E) versus B6_G5_R89,yes (D) do not connect but p-value is 0.9999.
Did you find others?
I am not sure what you mean by "but the Tukey table from JMP doesn't match with anything." Would you also post a picture of the corresponding SAS output to clarify?
This matter might need the attention of JMP Technical Support (support@jmp.com).
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Letters report vs differences report. Are they different?
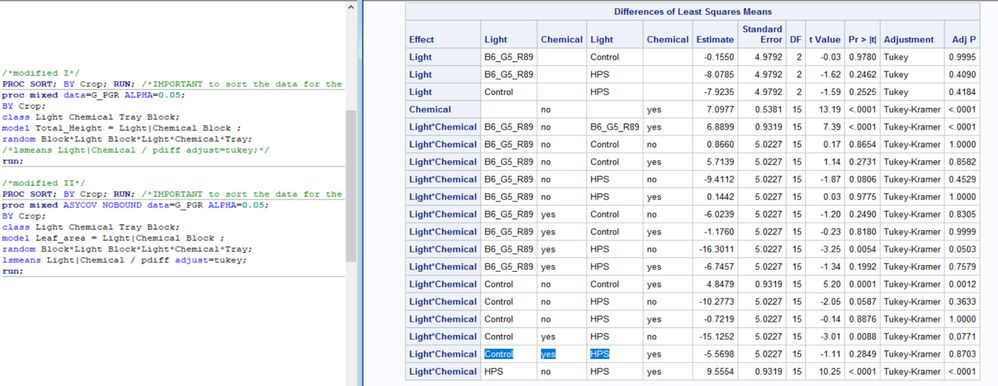
Sure, here is the SAS table and model (split-plot with subsamples and blocks).
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Letters report vs differences report. Are they different?
Thanks for your help Mark!
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Letters report vs differences report. Are they different?
Hi @Cristian,
Welcome to the community!
I only spot checked a few of these, but I don't see any mismatches. The connecting letters report is constructed such that any levels that share a level do not have a statistically significant difference. Since there are only 3 comparisons that are statistically significant in the table, it's easy to verify that those match. For example "HPS,no" vs. "HPS,yes" has a p-value < 0.001. In the connecting letters report, "HPS,no" has letters A D E, and "HPS,yes" has letters "B C". Since none of those letters match, the table and letters are consistent. Same follows for "B6_G5_R89,no" vs "B6_G5_R89,yes" and "Control,no" vs. "Control,yes".
I have to admit this is the most unusual looking connecting letters report I have seen. The reason is the std. error for all of the other comparisons is 5.023. Notice that if the compared levels have the same chemical, the standard error is only 0.932? How is your data structured? Is this a split plot experiment by chance?
*Edit: Good eye Mark on spotting those inconsistencies.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Letters report vs differences report. Are they different?
Hi Cameron,
Yes, "Yes" and "No" are different. I also understand why this is weird and how difficult this is going to be to explain that there is more power in the split-plot than in the main plot. But where I need help to understand is with the interpretation of the different JMP outputs because the ANOVA says that there is an interaction between ''Light'' (control, hps, and B6) and ''Chemical'' (yes/no) and the differences report only shows differences between "yes" and "no".
In additon, I understand from the table that there are differences between B6-yes (D), and control-yes (C,E) and HPS-yes (B,C) which would make more sense with the interaction in the ANOVA. The table says " levels not connected by the same letter are significantly different". Do I have a wrong interpretation of this table?
Thanks,
Cristian
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Letters report vs differences report. Are they different?
Thanks for sharing the SAS output along with the JMP output.
I confirmed that the two sets of results are the same but please note that:
- the order of the listing is not the same
- some estimates are of opposite sign, but that difference does not affect significance
- JMP reports the adjusted p-value (for multiple comparisons) but labels it as p-value, whereas SAS separately reports both the p-value and the adjusted p-value.
There is, though, as you noted and I previously confirmed, a discrepancy in the JMP results between the p-value and the connecting letters report for two pairs.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Letters report vs differences report. Are they different?
What version of JMP are you using? Thanks!
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Letters report vs differences report. Are they different?
Mark,
The JMP version is Pro 13.0.0.
Yes, my biggest issue is with the table (differences by letters) because If that table is wrong I'm going to need to re check all my new and old results. I dont want to think about it.
Altough the differences report dosen't show and interaction Between Lights and Chemicals and the ANOVA says that there is an interaction, in this case the different reports should be more correct because the ANOVA dosen't show an interaction if I use transformed data (Log). Does this make sense?
Thanks very much,
Cristian
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Letters report vs differences report. Are they different?
JMP separates information for you. The ANOVA is about the whole model. The Effect Tests is about each term in the model. The Parameter Estimates is about each level in each term. The differences report that uses connecting letters is not to be used in JMP Pro 13.0, but the other reports have no problems.
Transformation of variables often results in a simplification of the model. Non-linearities and even some interactions might no longer be necessary after some transformations are used.
Recommended Articles
- © 2026 JMP Statistical Discovery LLC. All Rights Reserved.
- Terms of Use
- Privacy Statement
- Contact Us