- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Discussions
Solve problems, and share tips and tricks with other JMP users.- JMP User Community
- :
- Discussions
- :
- Re: Choosing the right Comparison of means
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Choosing the right Comparison of means
Hello,
I'm analyzing biological data where the output is a continuous variable. The tests consists of different treatments on plants which are then assessed for a level of disease (my output). If I plot a graph, all I get is a bunch of means with very large error bars that overlap each other. So, I'm trying to do the analysis with the little letters, to confidently be able to say which treatments are different from each other.
Now the issue I have, is that each repeat consists of two data points which are not independent from each other (two parts of the same plant). In this case I heard that usual comparisons like Tukey-Kramer don't apply.
So my question is, which comparison should I use, or which data transformation, to make sure I'm doing things right?
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Choosing the right Comparison of means
It might be easier to understand the problem with an example table.
It sounds like you have data like this
Three or more treatments.
Multiple plants per treatment
Multiple sample points (tissue types) per plant
Multiple samples of each tissue
In the end, you might want to see the effect of the treatment on both leaf and stem for example for each treatment.
If the multiple treatments are something like control and several dilutions of the goo, then that would change some things too.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Choosing the right Comparison of means
In addition to everything my former colleague (I'm retired) @Byron_JMP asked, is the arrangement/combination of treatments and plants some sort of designed experiment or is it just happenstance treatments applied willy nilly to plants as nature or someone decided to administer? This could have a large impact on how you should optimally analyze the responses...and not just multiple comparison tests.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Choosing the right Comparison of means
@P_Bartell , I'm sure its a well-designed, well-controlled experiment. Plants are painfully slow, so planning is huge.
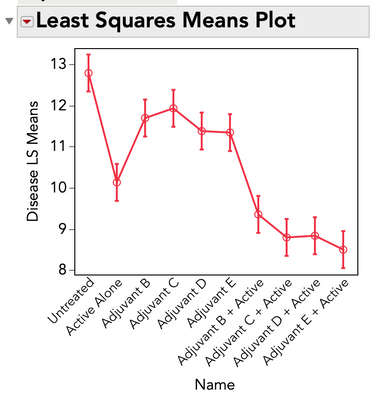
It's probably a multiple-variable problem, so a simple Turkey Kramer won't work. Just setting up the problem in Fit Model will help, and then we can look at the least squares means for the main treatment variable.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Choosing the right Comparison of means
Hello,
I like to believe it's well designed, cause I took part in the design process ;)
but jokes appart, it's plant especially grown for the test, with treatments applied to them in a standard, professional way.
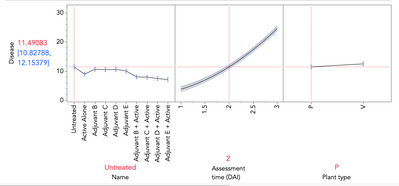
So, i got various treatments applied to 2 types of plants (variations of the same crop), and the level of disease (my output) assessed at different times (1,2,3 days). So we need to at least group things by assessment time.
Here's the data in anonimysed way, with the scripts attached. I look at both Student and Tukey tests, since one is deemed too permissive and the other a bit too restrictive. I don't see much differenciation here.
By the way gents, it's quite a bit of a pain to generate these connecting letters report in the fit Y by X tool. because I have 6 groups, that's 6 red triangles to click for the compare means. It's needlessly tedious. Do you jnow of a method to make things quicker?
ANyone reading this : have a look at this wish from the wishlsit and give it a kudo ! like this we could get the connecting letters built-into the graph builder.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Choosing the right Comparison of means
@P_Bartell @Byron_JMP , data table attached
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Choosing the right Comparison of means
Making sure I understand your data @bobmorrane : is each set of six rows one "subject" (what you are calling "Rep")? And then are the first three rows from each "subject" three repeated measures on one part of the plant, and the next three rows from each subject repeated measures on another part of the plant? And "Name" contains the treatments of interest (that you want to compare wrt their effects on those two parts of the plant, respectively). Do I have all of this correct?
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Choosing the right Comparison of means
Hi @MRB3855 ,
reps, or "repeats", are the same measurements done on different plants. So, plant 1, plant 2, plant 3. Each plant is grown separately in its own pot. for each rep, there are two measurements, which are done on two leaves of the same plant.
Then there's the assessment time. So, one measruement of the output value (disease) done on day one, then another the next day, and another one on day 3. Datapoints generated on the same day can be compared, but not over several days.
Name contains the treatment of interest yes.
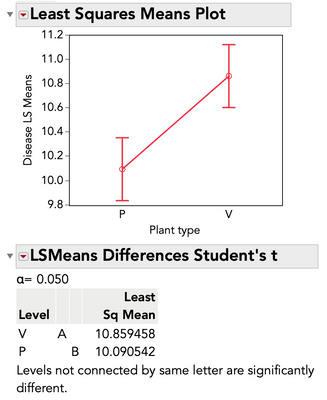
"Plant type" are two slightly different types of plants. The goal is to see whether one type is more resistant to the disease than the other. So, interesting to compare the disease value over both types of plant overall, or both types on a single treatment.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Choosing the right Comparison of means
Dude!!, this is such a great data set, wow.
I changed a couple column's modeling types and added a couple of new scripts to the table.
I tried modeling the data with a Response Surface Method (RSM). Rep(order?) seems to have an effect, so in model 2 I made that a Random Effect.
I'm not sure what you were hoping to find, but this is very interpretable data, and the study design is just fantastic too.
Fit Model(
Y( :Disease ),
Effects(
:Name, :"Assessment time (DAI)"n & RS, :Plant type,
:Name * :"Assessment time (DAI)"n,
:"Assessment time (DAI)"n * :"Assessment time (DAI)"n, :Name * :Plant type,
:"Assessment time (DAI)"n * :Plant type
),
Random Effects( :"Rep (Order)"n ),
Personality( "Standard Least Squares" ),
Emphasis( "Effect Leverage" ),
Method( "REML" )
);I changed the DAI column to numeric continuous, and the Rep Column to Numeric Continuous.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Choosing the right Comparison of means
Bob, I am not an SME regarding your situation, but I have looked at your table and have the following thoughts. Some of my questions and comments are more Socratic rather than require an answer.
1. First and foremost, is the variation in your response of any practical value? There is no context provided regarding how much of a change in disease is of scientific interest? This is always more important than statistical significance.
2. If I understand your data, it looks like you have one factor (Name) and it is tested at 10 "levels". You have 2 Plant types (crossed with Name). You have 10 plants (Rep) for each level of Name and each plant is measured twice (is this in different locations on the plant or the exact same location on the plant?) and measures of Disease over 3 time periods for "within" plant (you might consider the plants nested within Type and Name). We could debate whether the systematic sampling of time periods is crossed with Name. I have some questions regarding this structure. The numbering scheme for Rep looks questionable. Can the same plant be given a different level of Name? If not, there should be no repeated Rep numbers.How many plants are actually in the study? It also looks like you have 2 measures of disease for each plant? I have attached another version of the data table. It is imperative the data table match how the data was actually collected or any analysis will be suspect.
What is the intent of the study? (e.g., Are you trying to pick the "winner" (e.g., best Name to reduce max disease?) or trying to understand what contributes to disease (growth))? Why 10 plants? Why 3 time periods, each a day apart? Are you interested in the rate of change of disease over time or just maximum disease or what? I guess the question is what do you hypothesize the Name will do to disease? Why?
3. Have you assessed the measurement system? Did you measure the disease multiple times on the same location on same plant at the same time period? As it appears now, the measurement errors are likely confounded with within plant?
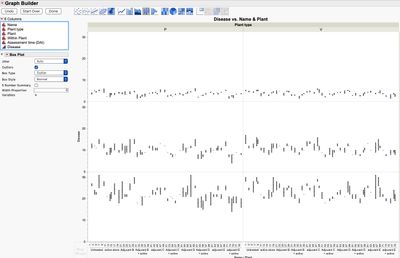
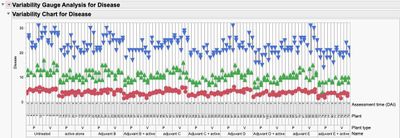
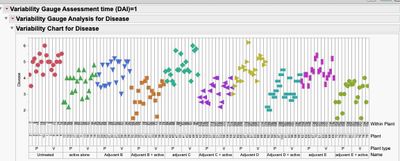
4. A graphical look at the data without any summarization shows the time periods (DAI) are the largest source of variation in the study. This may be expected and may be of no interest (hence why you may want to look at the data by DAI), but also notice the variation increases with DAI. There may also be some "outliers" in your data.
Colored by DAI
Colored by Name for DAI=1
Recommended Articles
- © 2026 JMP Statistical Discovery LLC. All Rights Reserved.
- Terms of Use
- Privacy Statement
- Contact Us