The add-in functions as a wrapper around the QuasR (short for Quantify and annotate short reads in R) package in BioConductor. It uses qAlign to align single or paired-end reads to a reference genome, and unmapped reads are optionally further aligned to alternative references. Pre-existing alignments in BAM format will be generated after importing.
The Experimental Design File is a tab-delimited text file or sas7bdat file containing the column names with "FileName" and "SampleName"; Raw sequence file can be compressed with gzip, bzip2 or xz (with extensions "gz", "bz2" or"xz"); A reference genome is required for creating the alignment index and further alignments.
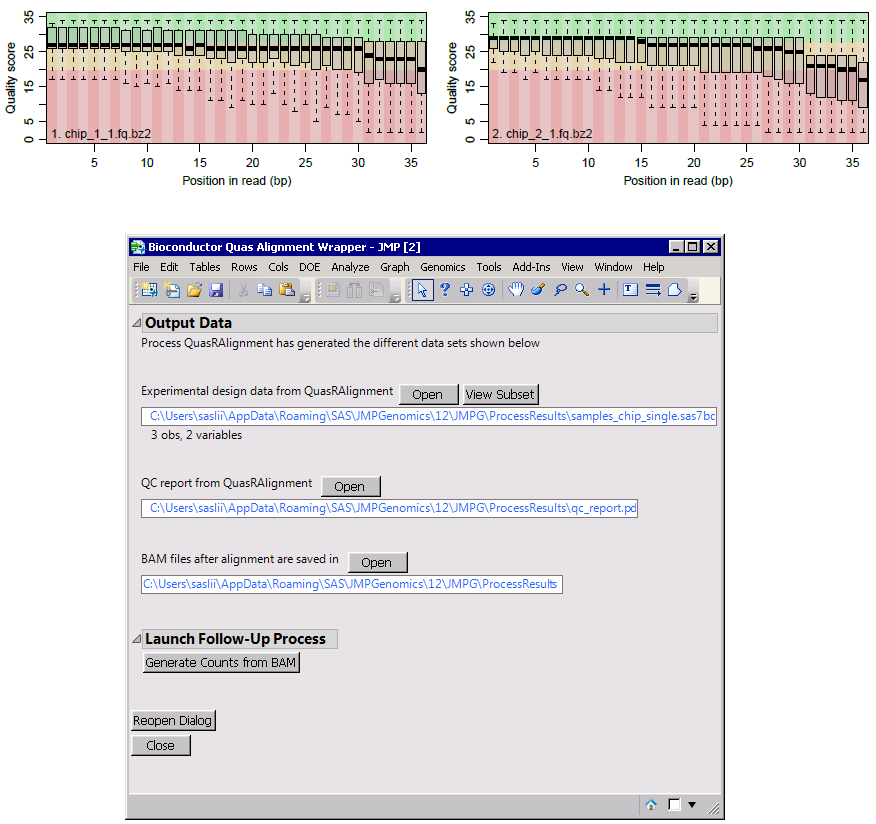
Output results: The alignments are generated and furhter analysis can be performed within JMP Genomics, for example, launching "Generate Counts from BAM" process. It also creates a quality control report with plots on sequence qualities and alignment statistics.
Citations:
Gaidatzis D, Lerch A, Hahne F, Stadler MB. QuasR: Quantification and annotation of short reads in R. Bioinformatics 31(7):1130-1132 (2015).
Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human
genome. Genome Biology 10(3):R25 (2009).
Au KF, Jiang H, Lin L, Xing Y, Wong WH. Detection of splice junctions from paired-end RNA-seq data by SpliceMap. Nucleic Acids Research, 38(14):4570-8 (2010).