- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Discussions
Solve problems, and share tips and tricks with other JMP users.- JMP User Community
- :
- Discussions
- :
- Re: multivariate data with a repeated measures design
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
multivariate data with a repeated measures design
Apologies for farming out what is more of a statistics than a JMP question to the community, but here goes: I was recently asked if JMP Pro could analyze a design in which 3,000 analytes are measured in the same individuals over time (one group perhaps receiving a medicine and the other having been given a placebo). The large number of analytes is not the problem, but the fact that this is a repeated-measures design and, to my knowledge, the multivariate options under Fit Model cannot handle a repeated measures design (would be no problem if there was a single Y). If I put in "time" into the model under MANOVA or partial least squares, that doesn't accommodate the repeated measures nature. Am I correct in assuming that the optimal statistical test I want cannot be performed in JMP Pro? Maybe instead I could look at "% change in concentration of each analyte" for each individual, thereby removing time from the model, but I am open to other options!
- Tags:
- macOS
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: multivariate data with a repeated measures design
I'm not a SME for your particular situation, but in general, first evaluate the within treatment variation (as quantified by your repeated measures). This might include graphically evaluating the repeats and assessing the consistency within treatment. Once you have evaluated the within treatment variation, you can decide how to enumerate those repeated measures (e.g., what statistics you want to use to summarize the within treatment data). This could be some measure of central tendency and some measure of variance. There is no one "right" way to do this. Your thought on using percentage change would certainly be worth trying. Then you analyze those summary statistics to model the treatment effects. The beauty of JMP is it will allow you to try multiple methods quite efficiently.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: multivariate data with a repeated measures design
Thank you for your thoughts. I think it may end up being simpler than I'm thinking because I have a feeling it will be presence-absence data: did the person develop a mutation, and, if so, which proteins changed in concentration over the period in which the mutation emerged? I think this is their question (outside of my area of expertise). In other words, it may not even need to be input into JMP as a typical repeated measures dialogue.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: multivariate data with a repeated measures design
Hi @abmayfield,
I may have another suggestion.
If your analyte response(s) are continuous values measured over time, could the Functional Data Explorer be helpful to extract the variation from the curves/ time evolution, with possible variance from repeated measurements for the same subject (ID) ?
There was a topic dealing with Measurement System Analysis for curve data that may be helpful in this context :
I hope this response will help you,
"It is not unusual for a well-designed experiment to analyze itself" (Box, Hunter and Hunter)
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: multivariate data with a repeated measures design
Thanks, and this is a cool idea with regard to using the FDE: did the biomarker should a "flatline" trend (i.e., no change), a hyperbolic one (shot up and went back down), or even a plateau (shot up and stayed up)? Ultimately, this may be what the physician running this clinical trial wants to know. I also suspect that JMP Clinical, which I have never used, LIKELY has tools to address just this sort of complex question, so I may explore that avenue, as well.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: multivariate data with a repeated measures design
Hi @abmayfield ,
JMP Clinical would not have anything additional to offer if looking for patterns over time except that it contains JMP Pro features like FDE (at least in JMP Clinical 17).
I think this is like chromatography data (amount of protein eluted over time) with a set of profiles (patients) for each analyze (protein). This could turn into what is Called FDE-DOE since a comparison of treatment is involved.
Maybe some helpful links:
In this case the DOE is really just a single factor of treated vs. untreated.
Data Scientist, Life Sciences - Global Technical Enablement
JMP Statistical Discovery, LLC. - Denver, CO
Tel: +1-919-531-9927 ▪ Mobile: +1-303-378-7419 ▪ E-mail: chris.kirchberg@jmp.com
www.jmp.com
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: multivariate data with a repeated measures design
I also like the FDE approach, but as a first step you might consider pivoting your data such that every value at every time is a different column, and there is one row per patient and then using PCA or PLS. With FDE you can easily/directly see the curve of each value over time, but you also need to perform the analysis for each y variable. With PLS the curves would be harder to interpret as they would not be automatically ordered by time or variable, but you would not need as many parameters.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: multivariate data with a repeated measures design
I actually may include this on the "wish list" because I think all of the elements are already there: PLS for multivariate analysis+mixed-model platform for repeated measures in which the repeated subject is directly specified. I think this is not as obscure or esoteric a need as it would seem because I think in the future, a lot of drug companies and clinical researchers will want to repeatedly profile entire suites of molecules in individuals tracked over time (who may or may not be taking a drug, for instance). I do feel, however, that doing this INcorrectly (i.e., ignoring the repeated-measures nature of the design) would NOT yield dramatically different results from the proper analysis assuming a large population of test subjects. In other words, if you gave a drug to 200 patients and a placebo to 200 others and then looked at gene expression changes over time with MANOVA or PLS, you could still detect differences even if people "started" at different places in terms of their baseline gene expression levels. But of course, doing it in the most statistically robust way would be preferable!
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: multivariate data with a repeated measures design
If you wanted to stick with the mixed model approach with repeated measures, I guess you could do PCA on the mRNA, save the largest impact PCs and then use those in the response role (kinda like what PLS will do). Then do the repeated measures as one might do in Mixed Models or Standard least squares like in this tutorial.
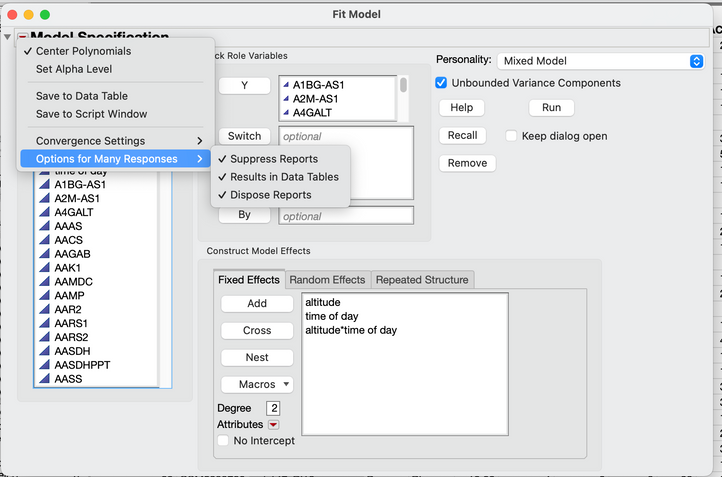
Or put all of the mRNA in response, choose mixed model, set it up as one would for a time based repeated structure and then put treatment in to Fixed Effects (I am probably missing something to add). Then use the red triangle and choose Options for Many Responses. You will get a table for everything instead of a report and that might help, but then there will be some extra work to sort out which mRNA is most affected and other stats for each of the model terms.
It's a thought anyway. Might be worth it.
Data Scientist, Life Sciences - Global Technical Enablement
JMP Statistical Discovery, LLC. - Denver, CO
Tel: +1-919-531-9927 ▪ Mobile: +1-303-378-7419 ▪ E-mail: chris.kirchberg@jmp.com
www.jmp.com
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: multivariate data with a repeated measures design
I was just going to come back in and reply with this, but you beat me to it. (I didn't see this reply before I replied yesterday, so I wonder if we were typing at the same time, @Chris_Kirchberg !)
This is a great option because you can then plot/sort the test p-values to see which are significant, etc. And with JMPs interactivity, you can select on the plots and it selects in the data table for subsetting out the "important" ones. This is how I've seen most 'omics-type analyses that have more complex random effects structures. Yes, it's ignoring the correlation between the Ys, but it makes the analysis feasible, whereas otherwise it's intractable.
Recommended Articles
- © 2026 JMP Statistical Discovery LLC. All Rights Reserved.
- Terms of Use
- Privacy Statement
- Contact Us