- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Discussions
Solve problems, and share tips and tricks with other JMP users.- JMP User Community
- :
- Discussions
- :
- Re: Single legend for Hierarchical clustering heat map
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Single legend for Hierarchical clustering heat map
Hi,
Thanks in advance for the support!
I'm making a Heatmap using 'two way clustering' in the hierarchical cluster. When I add the legend to the figure, it gives me one legend per column. I would like to have only one legend (color code) for the whole heatmap. I appreciate your guidance on that.
Shahram
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Single legend for Hierarchical clustering heat map
Thanks guys for your support!
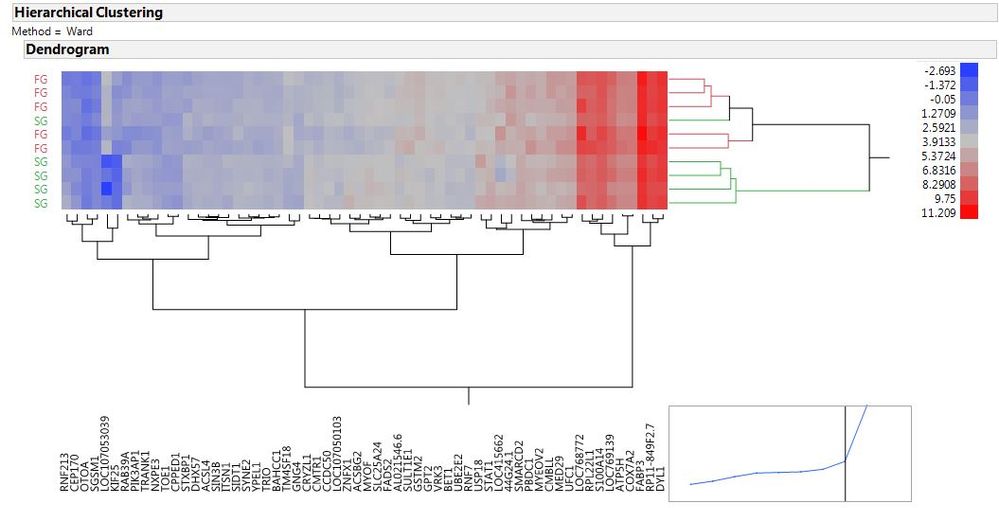
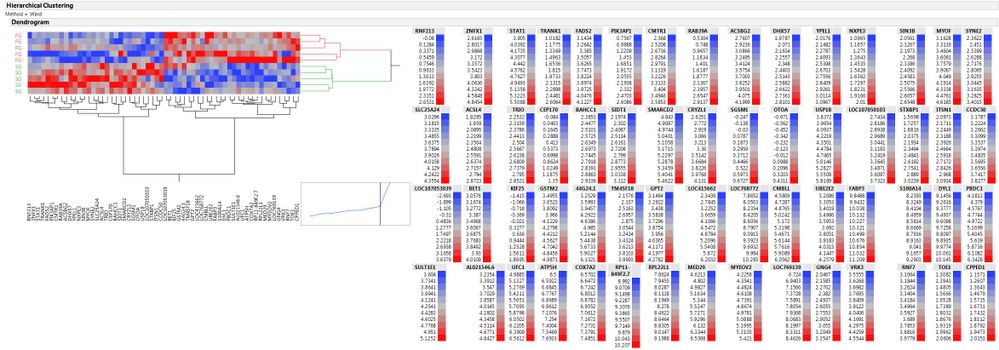
I have attached both pictures (standardized and non-standardized) to see the differences.
I'm using the JMP 13.2.1 (32-bit).
Jeff,
That's true each legend is a color code for each column which make sense.
Maybe I just use the legend of the non-standardized figure with the figure itself from standardized one together.
In that case, I get the figure that I want plus a single legend which indicates all the cell colors.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Single legend for Hierarchical clustering heat map
Thanks guys for your support!
I have attached both figures (standardized and non-standardized) to see the differences between them.
I'm using JMP 13.2.1 (32-bit).
Jeff, That's true. Each legend is a color code for each column. So if I want a standardized figure, I can't get a single legend.
Maybe, I just use the standardized heatmap with the legend from the non-standardized heatmap together. In that case, I will have the figure that I want with a single legend which contains the whole range of the color codes. It might not look proffessional though, but I think that's what people do!
Thanks.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Single legend for Hierarchical clustering heat map
@Jeff_Perkinson what does the lower graph show in the above cluster. I could not find the explanation.
Thanks in advance.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Single legend for Hierarchical clustering heat map
Hi,
To make a quick note about this graphic posted. The data used is already standardized (fold change) and thus was probably made by turning off the Standardize Data. All of the columns used for "Y" and are now on the top (or bottom) of the heatmap have the same units of measuremeant and mean the same thing (fold change) so they are directly compariable.
Do you have something similar where each column is in the same units and range of measurement?
Chris
Data Scientist, Life Sciences - Global Technical Enablement
JMP Statistical Discovery, LLC. - Denver, CO
Tel: +1-919-531-9927 ▪ Mobile: +1-303-378-7419 ▪ E-mail: chris.kirchberg@jmp.com
www.jmp.com
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Single legend for Hierarchical clustering heat map
Thanks Chris
The data that are used to make the graphs is RNA-seq results. There are two experimental conditions (FG and SG) which are the two horizontal clusters. And there are two main vertical clusters of each consist of 30 genes (extreme up- and down-regulated). Each cell should contain the expression level of each gene in one individual/sample (There are 10 samples here: 5 FG + 5 SG). The cell content/expression are all in FPKM which is already a normalized expression level and can be compared across genes and samples. Therefore, unit is the same for all the cell I guess. However, I can't understand how we can put the 'Fold Change' in the those cells. As far as I know, Fold Change is calculated by comparing two things. I'm not sure how Fold Change could be calculated for expression of ONE gene in ONE sample.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Single legend for Hierarchical clustering heat map
Thanks for the response. Sorry for the confusion, I was refering to the graphic from the journal you shared for fold changes.
You essentially have standarized data already. Standardize Data takes each Y (Column) and subtracts the column mean and divides by the column standard deviation (see below link).
http://www.jmp.com/support/help/13-2/Transformations_to_Y_Columns_Variables.shtml
Since you have normalized RNA-seq results in the same units, then definately uncheck standardize data since they all have the same scale (meaning) per sample for each gene (Y Column) . I am guessing that the normalized FPKM values are also log2 tranformed? You would then have over and under represented genes displayed as postive and negative, respectively, on the scale
I hope that helps.
Chris.
Data Scientist, Life Sciences - Global Technical Enablement
JMP Statistical Discovery, LLC. - Denver, CO
Tel: +1-919-531-9927 ▪ Mobile: +1-303-378-7419 ▪ E-mail: chris.kirchberg@jmp.com
www.jmp.com
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Single legend for Hierarchical clustering heat map
hi Chris,
I have a follow up question. If I already know the cluster results, say, using other clustering methods.
Can I generate a two-way cluster plot in JMP?
I guess it will be a simplified version, without the tree breaks to each column/rows, rather trucks into clusters.
Let me know if you have a quick advice for this?
Thanks very much.
Regards
Caroline
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Single legend for Hierarchical clustering heat map
HI, has this been resolved? I have the same question and could not find the solution in the thread.
Cheers,
Andreas
- « Previous
-
- 1
- 2
- Next »
Recommended Articles
- © 2026 JMP Statistical Discovery LLC. All Rights Reserved.
- Terms of Use
- Privacy Statement
- Contact Us