- JMP will suspend normal business operations for our Winter Holiday beginning on Wednesday, Dec. 24, 2025, at 5:00 p.m. ET (2:00 p.m. ET for JMP Accounts Receivable).
Regular business hours will resume at 9:00 a.m. EST on Friday, Jan. 2, 2026. - We’re retiring the File Exchange at the end of this year. The JMP Marketplace is now your destination for add-ins and extensions.
- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Discussions
Solve problems, and share tips and tricks with other JMP users.- JMP User Community
- :
- Discussions
- :
- Repeated vs Replicates vs multiple observations and nesting
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Repeated vs Replicates vs multiple observations and nesting
Hi JMP community,
I have been working on a biological data set where I need help in which model should I use to analyze my data. There are 2 different strains of mice (2 mice per strain) and 1 leg of each mouse has been given treatment 1, the other leg of the same mouse has been given treatment 2. I have taken 2-5 measurements during my experiment (chemical composition) per leg of each mouse. I am interested in using mixed model to compare means of chemical composition between each strain within a treatment and between treatment within a strain of mouse. Here is an example JMP table date set.
I don't particularly want to average the multiple observations, for the sake of making use of the in-sample variability in my stats. Thus, looking for a method where I can compare means, without losing the information that I get from making multiple observations, but also don't want to consider each observation as an independent observations or sample.
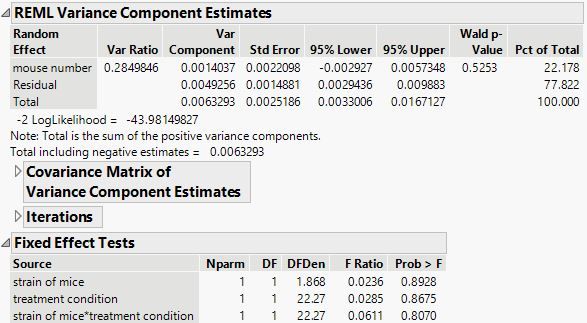
I use this particular table, and go to fit model> mixed model> then have mineral amount as a y variable, fixed effect as strain, treatment condition, strain* treatment condition, random effect as mouse number. I don't do any nesting and repeated measures. Should I be considering any?
Thank you for your help!
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Repeated vs Replicates vs multiple observations and nesting
What is the nature of the repeated measurements? Why did you measure the chemical more than once? Are the measurements taken close in time? Are the measurements taken over a period of time? Is time recorded so that it might be modeled and tested as another fixed effect?
Your model makes sense and using the individual measures makes sense, depending on how you answer these questions.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Repeated vs Replicates vs multiple observations and nesting
What is the nature of the repeated measurements? Why did you measure the chemical more than once? - measurement has been taken more than once, since it's a microscale imaging technique, which is very sensitive to sample surface roughness of the bone (extracted after sacrificing the mice). To avoid errors arising from the in sample variation, I did multiple measures, so I have a distribution of values to compare between samples.
Are the measurements taken close in time? Are the measurements taken over a period of time? Is time recorded so that it might be modeled and tested as another fixed effect? - All of the multiple measurements are not time sensitive, they are only 1 minute apart (the amount of time it takes the instrument to take a single measurement and move to next point on the sample surface, which is less than 10 microns away).
I guess, the two biggest concerns I have are that:
1) If I do mixed model, how do I make sure that the model is not considering every single measurement as a single independent sample?
2) How to I make sure that I am accounting for the fact that measures for treatment 1 and treatment 2 are coming from the same mouse?
Thank you!
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Repeated vs Replicates vs multiple observations and nesting
Thank you for answering my questions!
- The mixed model that you propose only assumes that every observation is sampling the same random error. It represents variation from sacrificing the mouse, harvesting the leg, and so on all the way to measurement error. That is to say, the errors are poolable into a single variance component. Your model is a statement that you do not expect the population of errors to vary mice. Another way to say it is that you are sampling the same population of errors regardless of which mouse.
- Well, mouse number acts like a blocking factor with a random effect so your proposed model should be correct. Additionally, the factor levels in your factor data columns should indicate the shared conditions. You factors are crossed, not nested. That is to day, Strain 1 is always Strain 1. Strain 1 is not something different in each mouse or treatment. Similarly, Treatment 1 is always the same treatment regardless of which mouse strain. Factors that are truly nested can be analyzed with JMP by indicating the specific nesting for each term in the model. Your measurements are blocked by mouse number.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Repeated vs Replicates vs multiple observations and nesting
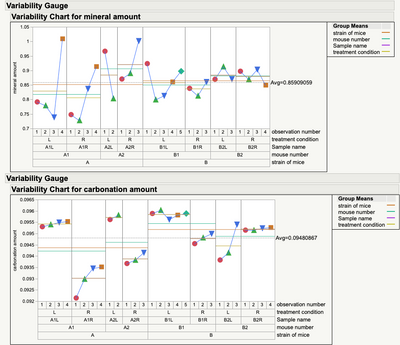
Only adding to what Mark has already said and given guidance on. The multiple measures taken in a short period of time are indeed nested and are a potentially good estimate of measurement error. I would first look at those measures. Do they make sense? A simple plot of the data will help. How much variation is there? Is the variation consistent? Are there any unusual measurements? Is an average a good estimate of those measures? (averaging will reduce the measurement error)
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Repeated vs Replicates vs multiple observations and nesting
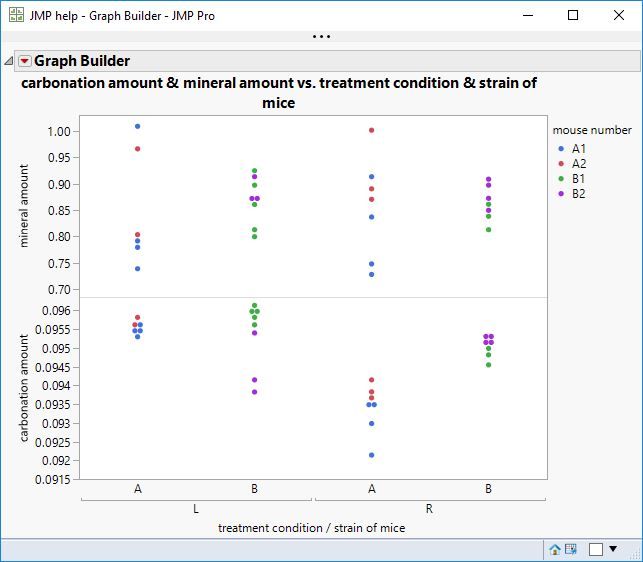
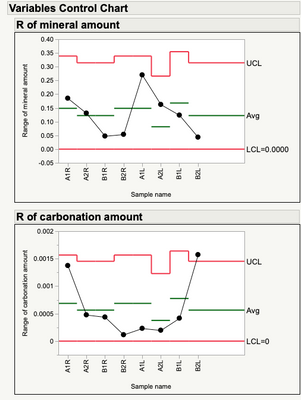
"You can see a lot just by looking."
As suggested by the plot, there is nothing to be found for the mineral amount.
The carbonation amount, on the other hand, shows some fixed effects.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Repeated vs Replicates vs multiple observations and nesting
You have some interesting measurement variation. Particularly where there are some "unusual data points" or special cause variability. Might want to look at those before going to far (analysis wise) with the averages.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Repeated vs Replicates vs multiple observations and nesting
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Repeated vs Replicates vs multiple observations and nesting
Thank you MarkBailey and Statman.
Yes, I have been thinking about the distribution of these multiple measurements, some of them were outliers (if the measurement comes from a natural asperity in the bone like a void, crack etc, then that can lead to outliers). This is not the actual/real data (apologize I forgot to mention earlier).
So what I have understood till now is:
1) Mixed model only assumes that the random error is uniform for all the samples
2) There is no nesting in this model since every mouse from each strain is receiving both treatments
3) The fact that the measurements for the two treatment conditions are dependent is being taken care of by designating mouse number as a random effect (blocking)
I have one last question:
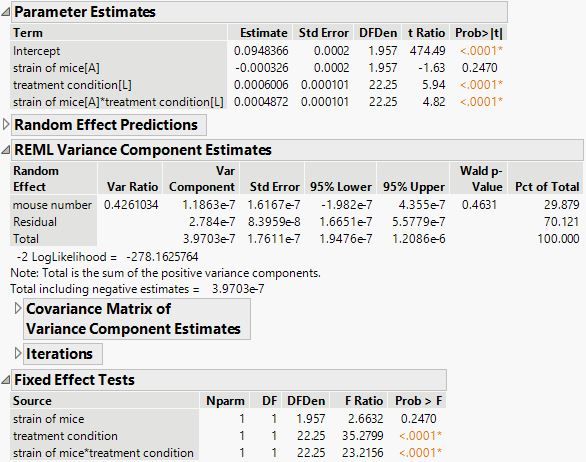
What would be the difference in running the tests for data with every single observation stacked versus, test for data where I take mean for every sample. Which method should I use?
Thank you!
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Repeated vs Replicates vs multiple observations and nesting
I'm not sure what you mean by stacked? The repeats data points can be analyzed for consistency and variability. Once you are satisfied with that you can choose how to represent those data points. As an average, standard deviation, variance, etc. or more the one summary statistic. You will need to summarize those to evaluate the other components of the study. They are not additional degrees of freedom.
Recommended Articles
- © 2026 JMP Statistical Discovery LLC. All Rights Reserved.
- Terms of Use
- Privacy Statement
- Contact Us