- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Discussions
Solve problems, and share tips and tricks with other JMP users.- JMP User Community
- :
- Discussions
- :
- Re: Problem with RSM fit for a factor
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Problem with RSM fit for a factor
Dear JMP professionals,
I was running a response-surface DoE with 5 factors (X_1 to X_5 in the attached dataset) and 5 responses (X_6 to X_10). I have severe outliers in the main response and a significant lack of fit. Now my questions:
1) How can I deal with such a situation, how can I still use this dataset (and response) to get some information from my DoE?
2) Can I exclude the outliers and still fit the RSM well?
3) Is there any data transformation, which might help to have better fits?
Thanks for your help!
Best
Accepted Solutions
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Problem with RSM fit for a factor
1. The transformation tries to find parameters that make the data more normally distributed.
2. The profiler and the saved prediction formula are in the original scale. Thats why changing the profiler to log scale makes the model more interpretable looking.
3. Well, when the data are super skewed, then the basic assumptions for a linear regression kind of get thrown out.
Also, and maybe more importantly, subject matter expertise informs the transformation. Something like, say, virus or cell counts tend to be log or log10 transformed. But since I don't know what you're measuring (wink/nod, by biologist friend), I just used the Box-Cox transformation JMP suggested, with an aim to get best model possible.
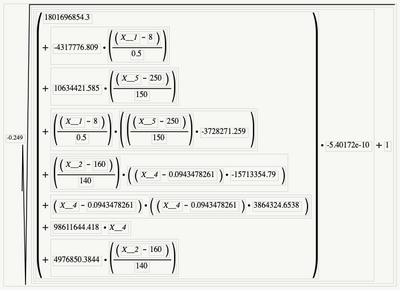
Prediction formula, back transforming the Box Cox transformation.
I'd like to take credit for the math, but JMP does it automatically
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Problem with RSM fit for a factor
"I have severe outliers in the main response and a significant lack of fit"
Which is your main response - X_6, X_7, X_8, X_9 or X_10?
What do you mean by "severe outliers"? I looked at residual plots for all 5 responses and only saw one observation that I would consider an obvious outlier: that was row 12 for response X_10 which is one of your centre points.
Concerning transformations, responses X_6 and X_8 are strongly skewed and will benefit from a Log transformation.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Problem with RSM fit for a factor
Hi David,
Thank a lot for your answer! The main response is X_6, X_7 and X_9. Especially X_6 is of main interest for the analysis.
I meant outliers because I have a strongly significant lack of fit (0.0019 and in the actual by predicted plot as well as in the residuals plot I see that the dataset is kind of "splitted" into to populations (see fit least square file below). All data below ~1 E+07 show a different tendency compared to the data above 1E+07 (here its is row 14,15, 17, 20 and 24). Even after clearing the dataset the lack of fit is still prominent and obvious in the actual by predicted plot and residuals by predicted plot.
I tried a log transformation of X_6 but the dataset does not look better or fits better (see log transformated file below).
How do I deal with this dataset and is the transformation here really the right one?
My hypothesis is, that this "splitted populations" are due to some nonlinearity of the device detector, which we used to measure the response (its not linear anymore above 1+E07. Is this an explaination for the dataset? Since the experiment was quite expensive and the results have big influence on our downstream process your help is highly appreciated. By the way, X_8 is a response, which I dont really need for my analysis.
Thanks a lot again for your time :) and help and I hope I can learn from you and the problems with the dataset for future projects.
Best regards,
Simon
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Problem with RSM fit for a factor
I messed around with modeling your data set a little.
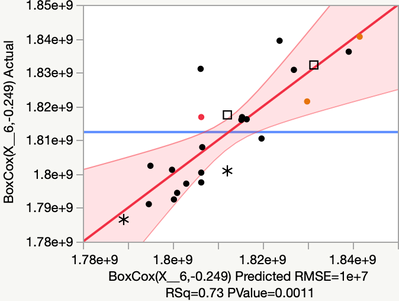
This model for X6 looks pretty good
Fit Model(
Y(
Transform Column(
"BoxCox(X__6,-0.249)",
Formula( (:X__6 ^ (-0.249) - 1) / -0.0000000005401720099 )
)
),
Effects(
:X__1 & RS, :X__5 & RS, :X__1 * :X__5, :X__2 * :X__4, :X__4 * :X__4, :X__4,
:X__2
),
Personality( "Standard Least Squares" ),
History( Y( :X__6 ) ),
Emphasis( "Effect Screening" ),
Run(
Profiler(
1,
Confidence Intervals( 1 ),
Term Value(
X__1( 8, Lock( 0 ), Show( 1 ) ),
X__5( 250, Lock( 0 ), Show( 1 ) ),
X__2( 160, Lock( 0 ), Show( 1 ) ),
X__4( 0.09292, Lock( 0 ), Show( 1 ) )
)
),
:"BoxCox(X__6,-0.249)"n << {Summary of Fit( 0 ), Analysis of Variance( 0 ),
Parameter Estimates( 1 ), Effect Details( 0 ), Sorted Estimates( 0 ),
Plot Actual by Predicted( 1 ), Plot Regression( 0 ),
Plot Residual by Predicted( 1 ), Plot Studentized Residuals( 1 ),
Plot Effect Leverage( 0 ), Plot Residual by Normal Quantiles( 0 ),
Box Cox Y Transformation( 1 )}
)
)There is a pretty wild looking transform for x6, but it was easy.
Turn on the Box Cox transformation option, then from the red triangle in the Box-Cox Transformations outline bar, choose Refit with Transform.
The model uses a reduced set of factors
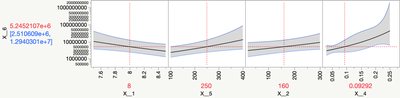
Also, try changing the Y-axis of the profiler to Log from Linear to see the response surface better.
Prediction Profiler
I might have cheated a little on getting the reduced model. In JMP Pro, I used Generalized Regression, Lasso, Leave One out validation, then relaunched with active effects and added back main effects (didn't enforce effect heredity in the lasso).
Excluding row 4 improves the model statistics. For x6 it looks like a potential outlier. After reducing the model, loosing this one run has a minimal effect on quality of the design.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Problem with RSM fit for a factor
Hi Bryon,
thanks lot for your answer and indeed the BoxCox transformation worked. Since I am just a biologist and quite new in the DoE world, I have a couple of questions here:
1) How does the BoxCox transformation effects the data reliability, since we do data transformation of the raw data?
2) The response values are transformed now, how can I see the untransformed responses when I change my factors in the prediction profiler. I need, that for comparability.
3) When is a data transformation suggested? It feels a bit arbitrary to me to just transform the dataset and than fit the data again, if the dataset is skewed.
Thanks a lot for your help and best wishes,
Simon
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Problem with RSM fit for a factor
1. The transformation tries to find parameters that make the data more normally distributed.
2. The profiler and the saved prediction formula are in the original scale. Thats why changing the profiler to log scale makes the model more interpretable looking.
3. Well, when the data are super skewed, then the basic assumptions for a linear regression kind of get thrown out.
Also, and maybe more importantly, subject matter expertise informs the transformation. Something like, say, virus or cell counts tend to be log or log10 transformed. But since I don't know what you're measuring (wink/nod, by biologist friend), I just used the Box-Cox transformation JMP suggested, with an aim to get best model possible.
Prediction formula, back transforming the Box Cox transformation.
I'd like to take credit for the math, but JMP does it automatically
Recommended Articles
- © 2026 JMP Statistical Discovery LLC. All Rights Reserved.
- Terms of Use
- Privacy Statement
- Contact Us