- Due to inclement weather, JMP support response times may be slower than usual during the week of January 26.
To submit a request for support, please send email to support@jmp.com.
We appreciate your patience at this time. - Register to see how to import and prepare Excel data on Jan. 30 from 2 to 3 p.m. ET.
- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Discussions
Solve problems, and share tips and tricks with other JMP users.- JMP User Community
- :
- Discussions
- :
- Re: JMP Genomics - P-value criteria for Basic RNA-Seq Workflow
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
JMP Genomics - P-value criteria for Basic RNA-Seq Workflow
When using the Basic RNA-Seq Workflow in JMP Genomics, how is significance defined?
Although I have specified the FDR p-value adjustment, the volcano plots still graph the unadjusted p values on the y-axis. Speaking of volcano plots, how is the horizontal line determined? This value seems to fluctuate between experiments, but I cannot find any indication of what it represents.
Under Output Data, it is not clear if the ANOVA results are filtered on the raw p-value or the adjusted p-values.
- Tags:
- windows
Accepted Solutions
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: JMP Genomics - P-value criteria for Basic RNA-Seq Workflow
Hi @LouisAltamura ,
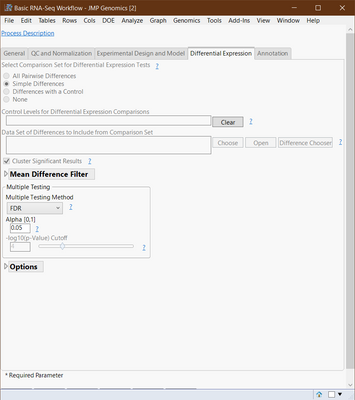
The un-adjusted -log10(p-values) are plotted on the y-axis of the volcano plot. The horizontal line drawn is the adjusted (in this case FDR adjusted) p-value cutoff defined in the Basic RNA-Seq Workflow dialog (see Multiple Testing section in the figure below). This cutoff is what is used to filter the transcripts so any transcript that has a raw -log10(p-value) greater or equal to this FDR adjusted cutoff is kept and the rest are filtered out.
Here is the online help regarding some more details the report output of the ANOVA results:
Hope this helps,
Data Scientist, Life Sciences - Global Technical Enablement
JMP Statistical Discovery, LLC. - Denver, CO
Tel: +1-919-531-9927 ▪ Mobile: +1-303-378-7419 ▪ E-mail: chris.kirchberg@jmp.com
www.jmp.com
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: JMP Genomics - P-value criteria for Basic RNA-Seq Workflow
Hi @LouisAltamura ,
The un-adjusted -log10(p-values) are plotted on the y-axis of the volcano plot. The horizontal line drawn is the adjusted (in this case FDR adjusted) p-value cutoff defined in the Basic RNA-Seq Workflow dialog (see Multiple Testing section in the figure below). This cutoff is what is used to filter the transcripts so any transcript that has a raw -log10(p-value) greater or equal to this FDR adjusted cutoff is kept and the rest are filtered out.
Here is the online help regarding some more details the report output of the ANOVA results:
Hope this helps,
Data Scientist, Life Sciences - Global Technical Enablement
JMP Statistical Discovery, LLC. - Denver, CO
Tel: +1-919-531-9927 ▪ Mobile: +1-303-378-7419 ▪ E-mail: chris.kirchberg@jmp.com
www.jmp.com
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: JMP Genomics - P-value criteria for Basic RNA-Seq Workflow
Thanks Chris! I am still using version 10.0 and the help dialog points to an older resource. This new user guide for 10.1 seems to be a lot clearer.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: JMP Genomics - P-value criteria for Basic RNA-Seq Workflow
Thanks @LouisAltamura ,
There is an update to 10.0. It is a hotfix that will bring your version to 10.1. You can check it out here:
https://www.jmp.com/support/notes/67/654.html
Glad I could help,
Data Scientist, Life Sciences - Global Technical Enablement
JMP Statistical Discovery, LLC. - Denver, CO
Tel: +1-919-531-9927 ▪ Mobile: +1-303-378-7419 ▪ E-mail: chris.kirchberg@jmp.com
www.jmp.com
Recommended Articles
- © 2026 JMP Statistical Discovery LLC. All Rights Reserved.
- Terms of Use
- Privacy Statement
- Contact Us