- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Discussions
Solve problems, and share tips and tricks with other JMP users.- JMP User Community
- :
- Discussions
- :
- How to record the classification of data points within ellipses in a Scatter Plo...
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
How to record the classification of data points within ellipses in a Scatter Plot Matrix after clustering?
The following JSL code can be used to display a ScatterPlot Matrix after Clustering.
dt = Open("$SAMPLE_DATA/Big Class.jmp");
Normal Mixtures(
Y( :height, :weight ),
{Mixtures Tolerance( 0.00000001 ), Mixtures MaxIter( 300 ),
Mixtures N Starts( 30 ), Outlier Cluster( 0 ), Diagonal Variance( 0 ),
Number of Clusters( 2 ), Go( Scatterplot Matrix( Ellipse Alpha( 0.1 ) ) )},
SendToReport(
Dispatch( {"Normal Mixtures NCluster=2"}, "Correlations for Normal Mixtures",
OutlineBox,
{Close( 0 )}
),
Dispatch( {"Normal Mixtures NCluster=2", "Scatterplot Matrix"}, "101",
ScaleBox,
{Min( 59.68 ), Max( 179.498666666667 ), Inc( 20 ), Minor Ticks( 0 )}
)
)
);
I need assistance with the following two questions:
(1) How can I set the Ellipse α to 0.4 in the JSL code?
(2) I would like to record the points included in the Ellipse with an α of 0.4, where the points within the red Ellipse are labeled as 1, those within the green Ellipse are labeled as -1, and the points outside both Ellipses are labeled as 0. This is similar to saving clusters in a Table, but instead, I want to record the points included in the Ellipses. How can I add this functionality to the JSL code?
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: How to record the classification of data points within ellipses in a Scatter Plot Matrix after clustering?
It seems like normal mixtures platform might be using it's own scatterplot matrix which isn't that easy to access or manipulate. You could save the clusters to your table and then use Scatterplot Matrix to create the plot
Scatterplot Matrix(
Y(:height, :weight),
Group(:Cluster),
Matrix Format("Lower Triangular"),
Density Ellipses(1),
Ellipses Coverage(0.4),
Group By(:Cluster)
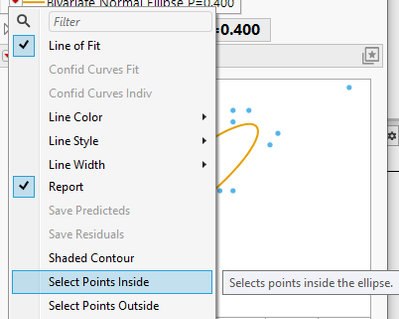
);but this won't let you get the points. To get this you can use Fit Y by X platform (use cluster as By column).
This is one place where I know you can select the points inside of the ellipses
As I'm not really sure what you are trying to do (are you just trying to select the points or also have some sort of a report) below is script which does a lot of things
Names Default To Here(1);
dt = Open("$SAMPLE_DATA/Big Class.jmp");
nw = New Window("",
vlb = V List Box(
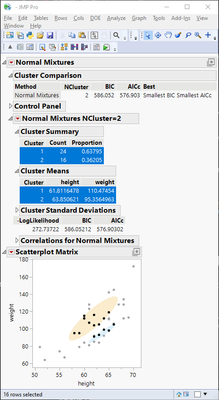
nm = dt << Normal Mixtures(
Y(:height, :weight),
{Mixtures Tolerance(0.00000001), Mixtures MaxIter(300), Mixtures N Starts(30),
Outlier Cluster(0), Diagonal Variance(0), Number of Clusters(2), Go()}
)
)
);
cols = Associative Array(dt << Get Column Names("String"));
nm << Save Clusters;
cluster_col = Associative Array(dt << Get Column Names("String"));
cluster_col << Remove(cols);
cluster_col = (cluster_col << get keys)[1];
vlb << Append(
sm = dt << Scatterplot Matrix(
Y(:height, :weight),
Group(:Cluster),
Matrix Format("Lower Triangular"),
Shaded Ellipses(1),
Ellipses Coverage(0.4),
Group By(Eval(cluster_col))
);
);
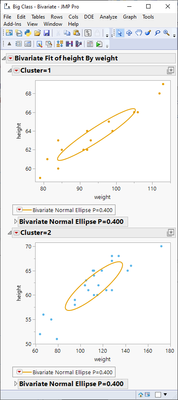
biv = dt << Bivariate(Y(:weight), X(:height), By(:Cluster), Invisible);
biv << Density Ellipse(0.4, {Select Points Inside});
new_col = dt << New Column("Ellipse", Numeric, Nominal, Formula(
If(Selected() & :Cluster == 1,
1
, Selected() & :Cluster == 2,
-1
,
0
)
));
dt << run formulas;
new_col << Delete Formula;
dt << clear select;
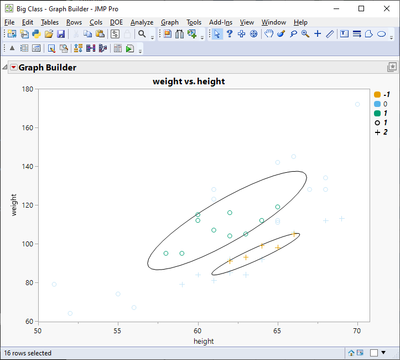
// Just for visualization purposes
gb = dt << Graph Builder(
Size(653, 491),
Show Control Panel(0),
Variables(
X(:height),
Y(:weight),
Overlay(:Cluster, Overlay Encoding("Style")),
Color(:Ellipse)
),
Elements(Points(X, Y, Legend(3)))
);
For(i = 1, i <= 2, i++,
ls = Report(biv[i])[FrameBox(1)] << Find Seg(LineSeg(1));
xs = ls << Get X Values;
ys = ls << Get y Values;
Eval(EvalExpr(
Report(gb)[FrameBox(1)] << Add Graphics Script(
Line(Expr(xs), Expr(ys));
)
));
);
//biv << close window;
Recommended Articles
- © 2026 JMP Statistical Discovery LLC. All Rights Reserved.
- Terms of Use
- Privacy Statement
- Contact Us