- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Discussions
Solve problems, and share tips and tricks with other JMP users.- JMP User Community
- :
- Discussions
- :
- Re: How to exclude rows until distribution mean meets a criteria?
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
How to exclude rows until distribution mean meets a criteria?
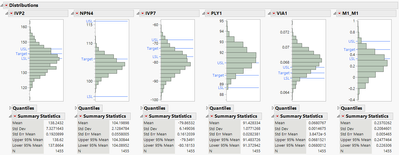
Below is an example similar to my actual case where I would like to exclude SITEs until the distribution mean for each parameter (only those plotted case below) fall within their respective spec limits.
Sites/wafer to be excluded start from the distribution extreme on the side where the mean is outside the spec limit (LSL or USL).
I think one could start with any parameter, remove sites from the distribution extreme until the recalculated mean is >=LSL or <= USL for that parameter, then move to the next parameter and check and exclude sites if necessary.
How to approach this problem with JSL?
Is there a JMP platform which could already do this, e.g. process screening?
The following gives
Names Default To Here( 1 );
Clear Log();
dt = Open( "$sample_data\Semiconductor Capability.jmp" );
obj = dt << Manage Spec Limits( Y( dt << Get Column Group( "Processes" ) ), Show Limits All, Save to Column Properties(1), );
obj << close window;
dist = Distribution(Stack( 0 ),
Continuous Distribution(Column( :IVP2 ), Outlier Box Plot( 0 ), Process Capability( 0 ) ),
Continuous Distribution(Column( :NPN4 ), Outlier Box Plot( 0 ), Process Capability( 0 ) ),
Continuous Distribution(Column( :IVP7 ), Outlier Box Plot( 0 ), Process Capability( 0 ) ),
Continuous Distribution(Column( :PLY1 ), Outlier Box Plot( 0 ), Process Capability( 0 ) ),
Continuous Distribution(Column( :VIA1 ), Outlier Box Plot( 0 ), Process Capability( 0 ) ),
Continuous Distribution(Column( :M1_M1 ), Outlier Box Plot( 0 ), Process Capability( 0 ) )
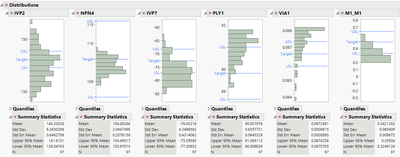
);while the desired end result may look something like (filtering done manually, plot below is when done for all wafers).
The act of filtering out SITEs using the recipe above is likely to throw the mean for earlier analysed parameters out of their spec limits. This is what I noticed while doing the exercise manually anyway. One may need loop the recipe say 5 times over the parameters for a check or until all SITEs have been filtered out. This is what I would like to know from this exercise. I would appreciate some help on how to proceed.
Once checked and done for all parameters, the excluded SITEs need to be tagged as Excluded in a new column (and remaining as Not Excluded).
- Tags:
- windows
Accepted Solutions
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: How to exclude rows until distribution mean meets a criteria?
Create expression/function to perform your exclusions. Then get list of columns of interest and loop over them one by one. I didn't verify this works correctly
Names Default To Here(1);
Clear Log();
dt = Open("$sample_data\Semiconductor Capability.jmp");
dt << clear row states();
obj = dt << Manage Spec Limits(
Y(dt << Get Column Group("Processes")),
Show Limits All,
Save to Column Properties(1)
);
obj << close window;
expr_exclude_rows = Expr(
Summarize(dt, colMean = Mean(dt:colName), colMin = Min(dt:colName), colMax = Max(dt:colName));
del_LSL = _LSL - colMean;
del_USL = _USL - colMean;
If(del_LSL > 0,
dt << select where(dt:colName == colMin) << Hide and Exclude;
);
If(del_USL < 0,
dt << select where(dt:colName == colMax) << Hide and Exclude;
);
If(N Items(dt << Get Selected Rows) < 1,
continue_clean = 0;
);
dt << Clear Select;
);
dt << Show Window(0);
dt << Begin Data Update;
duration = 0;
cols_of_interest = {"IVP7"};
For Each({colName}, cols_of_interest,
start = Tick Seconds();
specs = dt:colName << get property("spec limits");
_LSL = specs["LSL"];
_USL = specs["USL"];
continue_clean = 1;
While(continue_clean,
expr_exclude_rows;
);
end = Tick Seconds();
cur_dur = end - start;
duration += cur_dur;
Write("colName: ", colName, " took ", Round(cur_dur, 2), " seconds.\!N");
);
dt << End Data Update;
dt << Show Window(1);
total_end = Tick Seconds();
rows_excluded = dt << Get Excluded Rows;
Write("Total run time for ", N Items(cols_of_interest), " columns was " , Round(duration, 2), " seconds with ", N items(rows_excluded), " rows excluded\!N");- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: How to exclude rows until distribution mean meets a criteria?
Here is an illustration of how to run this separately for each wafer. I am using @jthi example code. I modified it to create data tables for each wafer, and then to run the code on each data table. For illustration purposes, I removed all wafer data except for wafers 1 & 2.
Names Default To Here( 1 );
Clear Log();
dtAll = Open( "$sample_data\Semiconductor Capability.jmp" );
dtAll << clear row states();
obj = dtAll << Manage Spec Limits(
Y( dtAll << Get Column Group( "Processes" ) ),
Show Limits All,
Save to Column Properties( 1 )
);
obj << close window;
Window( "Consistency Problem" ) << close window;
// For illustration remove all data except for wafers 1 & 2
dtAll << select where( :wafer > 2 );
dtAll << delete rows;
expr_exclude_rows = Expr(
Summarize( dt, colMean = Mean( dt:colName ), colMin = Min( dt:colName ), colMax = Max( dt:colName ) );
del_LSL = _LSL - colMean;
del_USL = _USL - colMean;
If( del_LSL > 0,
dt << select where( dt:colName == colMin ) << Hide and Exclude
);
If( del_USL < 0,
dt << select where( dt:colName == colMax ) << Hide and Exclude
);
If( N Items( dt << Get Selected Rows ) < 1,
continue_clean = 0
);
dt << Clear Select;
);
// Create all data tables based on wafer
dtList = dtAll << Subset( By( :wafer ), All rows, Selected columns only( 0 ) );
// Run the analysis for each data table
For Each( {dt}, dtList,
dt << Show Window( 0 );
dt << Begin Data Update;
duration = 0;
cols_of_interest = {"IVP7"};
For Each( {colName}, cols_of_interest,
start = Tick Seconds();
specs = dt:colName << get property( "spec limits" );
_LSL = specs["LSL"];
_USL = specs["USL"];
continue_clean = 1;
While( continue_clean, expr_exclude_rows );
end = Tick Seconds();
cur_dur = end - start;
duration += cur_dur;
Write( "colName: ", colName, " took ", Round( cur_dur, 2 ), " seconds.\!N" );
);
dt << End Data Update;
dt << Show Window( 1 );
total_end = Tick Seconds();
rows_excluded = dt << Get Excluded Rows;
Write(
"Total run time for ",
N Items( cols_of_interest ),
" columns was ",
Round( duration, 2 ),
" seconds with ",
N Items( rows_excluded ),
" rows excluded\!N"
);
);- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: How to exclude rows until distribution mean meets a criteria?
Hi @Neo,
Is your goal to specifically get those means inside the control limits, or are you looking for an automated way to clean up your dataset? If the latter, you might consider the Explore Outliers platform under the Analyze > Screening menu. I frequently use the quantile range outliers, and the Robust PCA outliers is very similar to what I typically do inside the PCA platform.
If you are specifically trying to find a set of sites (there are only 5?) or rows that satisfy the condition where they are all within the mean, then I would probably start with a multivariate analysis in order to find rows that have extreme values in multiple columns first.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: How to exclude rows until distribution mean meets a criteria?
Goal is to get the Mean inside Spec Limits for all concerned parameters and not to explore outliers.
Yes, in this example there are 5 sites per lot/wafer (>1000 in my actual case) but I could not find a better example in JMPs sample data.
Had a quick look at Multivariate analysis. I need help with more specifics I should look for under this platform.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: How to exclude rows until distribution mean meets a criteria?
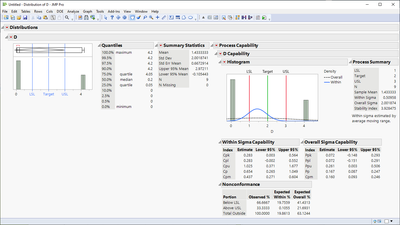
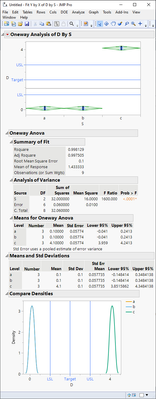
Are you ok with situations like this? Mean is still between the spec limits even though there isn't single result within the spec
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: How to exclude rows until distribution mean meets a criteria?
@jthi Thanks for pointing out one of the possible cases which satisfies the requirement but not the objective. I do not understand the Oneway analysis chart but the answer is No for the Distribution chart. In my example, I have got rows within the distribution but you are correct to point out a possibility.
Goal is to bring the Mean of a distribution inside Spec Limits by filtering out (Sites/Wafer in my example case) starting from the extremes of the distribution. While doing this manually, I excluded rows (sites) starting at the distribution extreme on the side where the Mean was outside the spec limit and as mentioned above I had to go back to previously analysed parameters as filtering out sites on subsequent parameters resulted in the mean of previously analysed parameters walking out of the spec limits.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: How to exclude rows until distribution mean meets a criteria?
@jthi Below is a skeleton dry run script which appears to work like I want it to. Its is only one chosen parameter now (will need to repeat for another parameters). I am struggling to get the steps under the correct loop.
I have steps in stages below (two shown below as commented text) which I need to run for each wafer until the if statement breaks out (there could be better way to do this e.g using While(), but this is what I have got to so far). I need help on the following
- How do I loop the tasks so that the min/max and mean per wafer is used for the recipe and not the corresponding values for entire parameter column.
- Once I have done this for one parameter column, I would like to move to the next (eg. total 6 in original post). How to do this automatically via JSL?
Names Default To Here( 1 );
Clear Log();
dt = Open( "$sample_data\Semiconductor Capability.jmp" ); // get data table
//get spec limits into column properties
obj = dt << Manage Spec Limits( Y( dt << Get Column Group( "Processes" ) ), Show Limits All, Save to Column Properties(1));
obj << close window; // close Manage Spec Limits window
dt << clear row states(); // clear any excluded or hidden rows
colName = "IVP7"; // choose column to operate on
specs = dt:colName << get property("spec limits"); //get spec limits for chosen column
_LSL = specs["LSL"]; //show (lsl); // get LSL
_USL = specs["USL"]; //show (usl); // get USL
/////// Stage 1 Start //////
colMean = Col Mean(dt:colName); show (colMean); // get col mean
colMin = Col Minimum(dt:colName); show (colMin); // get col min
colMax = Col Maximum (dt:colName); show (colMax); // get col max
del_LSL = _LSL - colMean; show (del_LSL); // get delta of mean wrt to LSL, +ve if colMean < LSL
del_USL = _USL - colMean; show (del_USL); // get delta of mean wrt to USL, -ve if colMean > USL
if (del_LSL >0, dt << select where( dt:colName == colMin)<< Hide and Exclude, // hide and exclude colMin
del_USL <0, dt << select where( dt:colName == colMax)<< Hide and Exclude, // hide and exclude colMax
break () // break if del_LSL<=0 or if del_USL >=0
);
/////// Stage 1 End/////
/////// Stage 2 Start //////
colMeanNew = Col Mean( If( Excluded(Row State(Row())), ., dt:colName ) ); show (colMeanNew);
colMinNew = Col Minimum( If( Excluded(Row State(Row())), ., dt:colName ) ); show (colMinNew);
colMaxNew = Col Maximum( If( Excluded(Row State(Row())), ., dt:colName ) ); show (colMaxNew);
del_LSL_New = _LSL - colMeanNew; show (del_LSL); // get new delta of mean wrt to LSL, +ve if colMean < LSL
del_USL_New = _USL - colMeanNew; show (del_USL); // get new delta of mean wrt to USL, -ve if colMean > USL
if (del_LSL_New >0, dt << select where( dt:colName == colMinNew)<< Hide and Exclude, // hide and exclude colMin
del_USL_New <0, dt << select where( dt:colName == colMaxNew)<< Hide and Exclude, // hide and exclude colMax
break () // break if del_LSL<=0 or if del_USL >=0
);
/////// Stage 2 End //////
/////// Stage 3 Start and so on//////
colMeanNew2 = Col Mean( If( Excluded( Row State(Row())), ., dt:colName ) ); show (colMeanNew2);
colMinNew2 = Col Minimum( If( Excluded( Row State(Row())), ., dt:colName ) ); show (colMinNew2);
colMaxNew2 = Col Maximum( If( Excluded( Row State(Row())), ., dt:colName ) ); show (colMaxNew2);
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: How to exclude rows until distribution mean meets a criteria?
Create expression/function to perform your exclusions. Then get list of columns of interest and loop over them one by one. I didn't verify this works correctly
Names Default To Here(1);
Clear Log();
dt = Open("$sample_data\Semiconductor Capability.jmp");
dt << clear row states();
obj = dt << Manage Spec Limits(
Y(dt << Get Column Group("Processes")),
Show Limits All,
Save to Column Properties(1)
);
obj << close window;
expr_exclude_rows = Expr(
Summarize(dt, colMean = Mean(dt:colName), colMin = Min(dt:colName), colMax = Max(dt:colName));
del_LSL = _LSL - colMean;
del_USL = _USL - colMean;
If(del_LSL > 0,
dt << select where(dt:colName == colMin) << Hide and Exclude;
);
If(del_USL < 0,
dt << select where(dt:colName == colMax) << Hide and Exclude;
);
If(N Items(dt << Get Selected Rows) < 1,
continue_clean = 0;
);
dt << Clear Select;
);
dt << Show Window(0);
dt << Begin Data Update;
duration = 0;
cols_of_interest = {"IVP7"};
For Each({colName}, cols_of_interest,
start = Tick Seconds();
specs = dt:colName << get property("spec limits");
_LSL = specs["LSL"];
_USL = specs["USL"];
continue_clean = 1;
While(continue_clean,
expr_exclude_rows;
);
end = Tick Seconds();
cur_dur = end - start;
duration += cur_dur;
Write("colName: ", colName, " took ", Round(cur_dur, 2), " seconds.\!N");
);
dt << End Data Update;
dt << Show Window(1);
total_end = Tick Seconds();
rows_excluded = dt << Get Excluded Rows;
Write("Total run time for ", N Items(cols_of_interest), " columns was " , Round(duration, 2), " seconds with ", N items(rows_excluded), " rows excluded\!N");- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: How to exclude rows until distribution mean meets a criteria?
@jthi Thanks. The key bit I was missing in my attempts at a While loop is
If(N Items(dt << Get Selected Rows) < 1,
continue_clean = 0;
);
dt << Clear Select;so it was not running as expected.
I have now tested it and modified it to work for several parameter columns following your script. Still some more testing needed but I think it works as required.
What I need to do next it repeat this analysis for every Wafer. Currently the script runs on all rows in the data table for the chosen parameter column. It is not clear to me on how to limit the analysis to one wafer before proceeding to the next.
End goal is to get the number of excluded rows per wafer (Perhaps adding an new column like ExcludedRows & set =1 in the data table as a part of the analysis could be useful)
But before I request some direction on how to script for repeating the analysis per Wafer.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: How to exclude rows until distribution mean meets a criteria?
Given the analysis you are performing, I believe the most efficient way of running for each wafer is to subset the data table into separate data tables for each wafer. Then you just step through the tables one after another,.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: How to exclude rows until distribution mean meets a criteria?
@txnelson Ok. Thanks, I know how subset by wafer (In my actual case, this will generate >100 wafer data) but I do not know how to script to run the analysis on wafer by wafer basis, once I have separate wafer data. Could I get some direction please?
Recommended Articles
- © 2026 JMP Statistical Discovery LLC. All Rights Reserved.
- Terms of Use
- Privacy Statement
- Contact Us