- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Discussions
Solve problems, and share tips and tricks with other JMP users.- JMP User Community
- :
- Discussions
- :
- Re: How to create a Nested effect within interaction factors?
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
How to create a Nested effect within interaction factors?
Hello,
I wil be running RCBD using fit model. I know how to create the nested effect within a single factor, but how do I create nested effect within interaction factors?
ex. Block (Year * Location)
Thank you!
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: How to create a Nested effect within interaction factors?
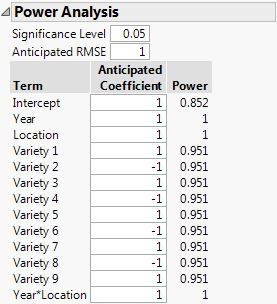
I don't know the standard deviation of your response or the minimum effect that you are trying to detect but this comparison is fair because I use 1 for the standard deviation (Anticipated RMSE) and 1 for the parameter value (with coding of factor levels, the value is equivalent to an effect that is twice the standard deviation) in both analyses. Here is the power analysis using the random block effect in the model:
(RCBD with random block effects in the model)
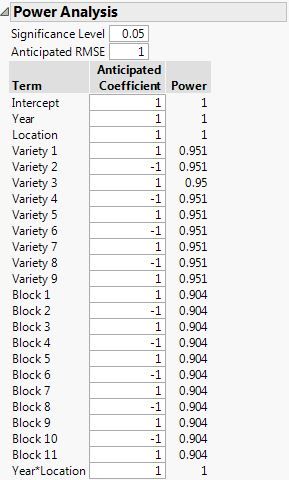
Here is the power analysis using your model:
(RCBD with fixed block effects in the model)
So you can see that the power is essentially the same for the effects of interest. Your choice should come down to how you think about the block effect. A random effect seems appropriate to me. Why would you want to model Block as a fixed effect? I do not mean that question as a challenge. It is simply something to think about.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: How to create a Nested effect within interaction factors?
Hello Mark,
Thank you for comparing the power of detection for me. I did't want to treat the nested Block (Year*Location) effect as fixed, effect. I wanted to treat it as random effect, but just didn't know how to run the test by nesting the Block within the interactions of Year and Location. But now since the power of detection was not affected by simply naming the Blocks 1~12 throughhout the 2 Locations and 2 Years, I would do the analysis this way. Thank you very cmuh again!!
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: How to create a Nested effect within interaction factors?
Dear Mark,
Thank you for this discussion of RCBD design 3 years ago. I followed this discussion, but I am still uncertain how to specify my model, which is actually much more simple. It also includes an effect nested within a combination of two other effects. Specifically, I have 4 genotypes reared at 2 temperatures and 4-5 individuals per combination of temperature and phenotype measured repeatedly (many observations of swimming speed per individual). So individuals are random effect nested within genotypeXtemperature. When I do as you suggested in the discussion above to specify individual with unique IDs and run this:
Fit Model( Y( :v ), Effects( :Genotype, :T, :T * :Genotype, :G_T_Ind & Random))
I get lost DF's and nothing is evaluated.
There must be a way to specify that individuals are unique within a combination of :T * :Genotype, right?
Thanks in advance!
L.Yampolsky
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: How to create a Nested effect within interaction factors?
Here is a simple example to illustrate how I interpret your design.
You included 4 x 2 = 8 treatments. Each treatment was replicated with multiple subjects (e.g., 5 times). Each subject was observed multiple times (e.g., 3 times). Is that essentially what you did?
So you could use the average of the observations as the response from each individual. This way, the residual error is individual and you do not need a term in the model. (You could also create and model the variance of the observations to see if the treatment affected the variability, too.)
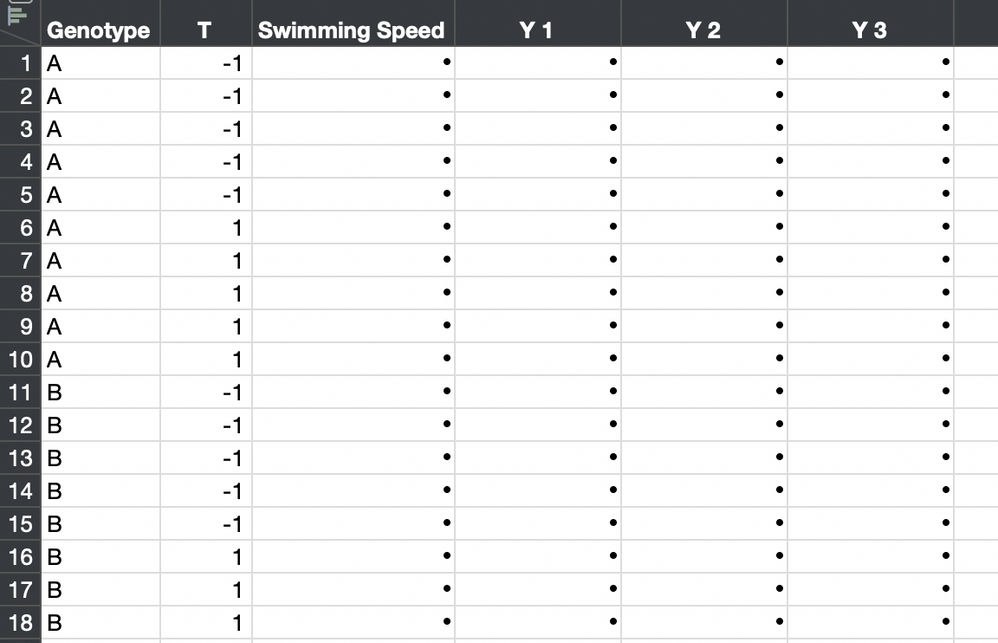
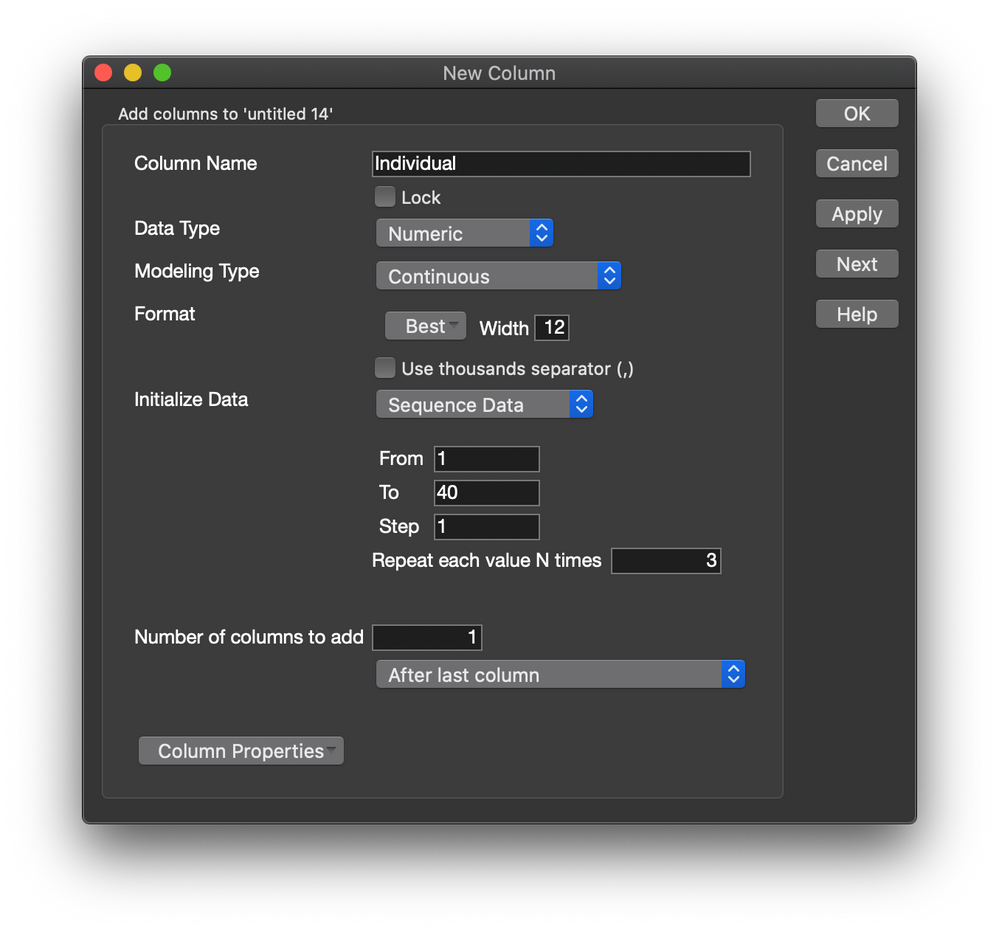
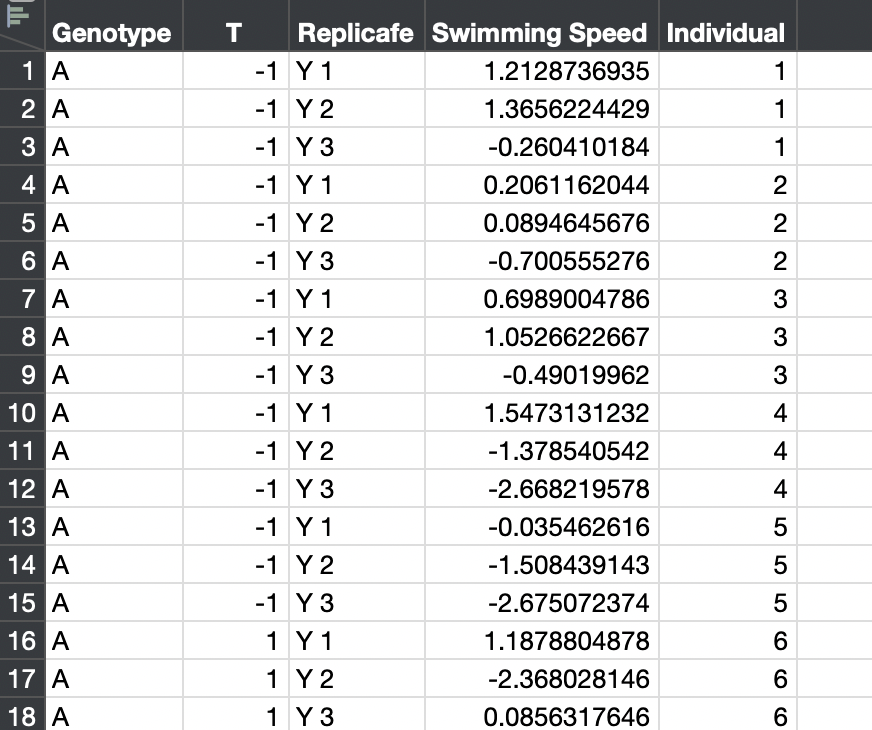
Alternatively, you could use the individual observations. You must stack the separate observation columns first and create a new column for the identity of the individual. The ID column would be added as the random effect.
The new table looks like this:
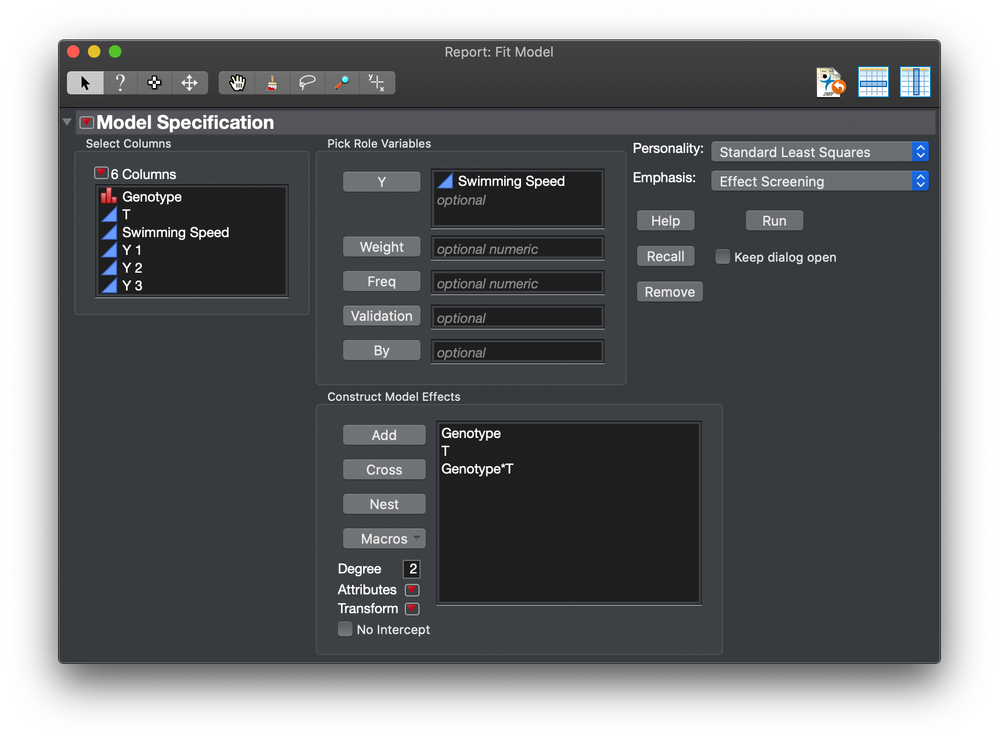
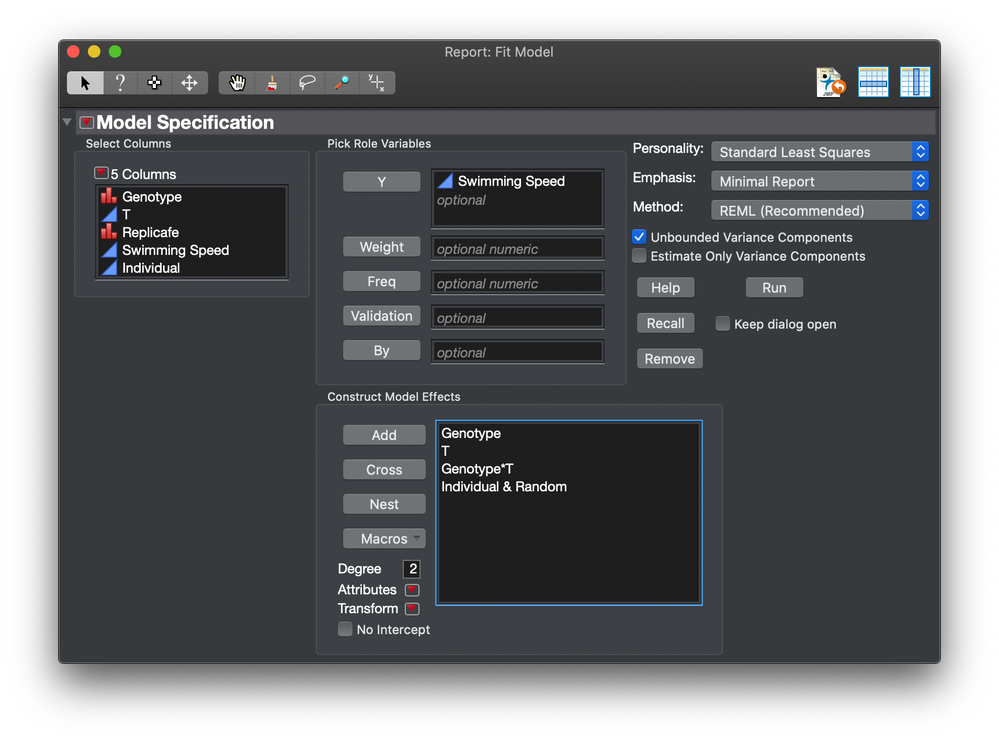
So the model changes to this:
This way, the individual contributes a random effect but it is not a nested effect. It acts like a random block.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: How to create a Nested effect within interaction factors?
Dear Mark,
Thank you so much, so quickly and on a weekend, you guys are amazing.
Yes this is pretty much my design, although each individual is measured not 3 times but more like 300 times (velocity recorded every second for 5 min and it varies quite a bit as individuals swim, rest, swim again, etc.). Of course these 300 measurements are not quite independent observations, but I feel they still can be used to estimate the difference between individuals. (Two adjacent measurement at not independent at all, but over a couple of cycles of swimming and resting perhaps they are).
Yes, sure, I started out by averaging speed for each individual and using individuals as replicates. But I would also like to be able to report that individuals within genotypes are quite different. So I'd like to use MS rather than REML method, because REML does not test random effects.
So when I did it exactly the way you show, with individuals given unique IDs, and genotype and T as full factorial effects and Individual as random non-interacting block effect. And I am getting a lot of singularities and missing DFs, as if JMP was trying to cross individuals to genotype and T, when using MS. (Everthing fine in REML).
Perhaps I cannot estimate the significance the Individual effect in the same model and should do it in a separate analysis, say for each T separately and nested within genotypes?
Thanks again for your advise!
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: How to create a Nested effect within interaction factors?
You said, "I would also like to be able to report that individuals within genotypes are quite different. So I'd like to use MS rather than REML method, because REML does not test random effects." The random effect is modeled as a variance. The REML report includes confidence intervals and a Wald test statistic. You can also examine the best linear unbiased prediction (BLUPs) for inidivuals.
You said, "So when I did it exactly the way you show, with individuals given unique IDs, and genotype and T as full factorial effects and Individual as random non-interacting block effect. And I am getting a lot of singularities and missing DFs, as if JMP was trying to cross individuals to genotype and T, when using MS. (Everthing fine in REML)." That is one of the advantages of REML. Many designs are not suited to the older MS method.
- « Previous
-
- 1
- 2
- Next »
Recommended Articles
- © 2026 JMP Statistical Discovery LLC. All Rights Reserved.
- Terms of Use
- Privacy Statement
- Contact Us