- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Discussions
Solve problems, and share tips and tricks with other JMP users.- JMP User Community
- :
- Discussions
- :
- Re: Friedman test on JMP?
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Friedman test on JMP?
Dear all,
I am fairly new in using the JMP software, and as far as I can see there is no function in the programme for using the Friedman test? I have a non-normal distribution of my different sets of values (more than three groups) and is opting for a non-parametric test. That is, I had like to test a 2-way type of test (as ANOVA). Wilcoxon test is insufficient since it is a one-way test.
Do anybody know how I will deal with is problem? Is there another test to perform instead of Friedman test?
Best regards,
Accepted Solutions
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Friedman test on JMP?
Hi Jim,
Friedman's test is not supported directly in JMP but, as Steve points out you can do this by calculating the ranks within each block and then doing a two way ANOVA on ranks using Fit Y by X (treating one of the effects as a blocking variable).
Computing the ranks within block is easy in JMP. Just use the Distribution platform with your original response as Y and your blocking variable as a By variable. Then, in the resulting report, hold down the Ctrl key and click on the little red triangle at the top of one of the histograms and choose Save->Ranks. You'll get a new column in your data table with the ranks within levels of your blocking variable.
I hope that helps!
-Jeff
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Friedman test on JMP?
If you can't find the Friedman test in the menus (and I have no idea whether you can or not, since I am not a JMP user), you could rank transform your data, and use a two-way ANOVA on the ranks. In SAS, this is available in PROC FREQ, so I would check the JMP documentation for the possibility of doing Friedman's test in that manner.
Steve Denham
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Friedman test on JMP?
Thanks for the help. Although the Friedman test seem not listed in the JMP menu or elsewhere in the programme. Perhaps I need to perform it on SAS if JMP doesnt conduct these tests.
Best regards,
Jim
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Friedman test on JMP?
Hi Jim,
Friedman's test is not supported directly in JMP but, as Steve points out you can do this by calculating the ranks within each block and then doing a two way ANOVA on ranks using Fit Y by X (treating one of the effects as a blocking variable).
Computing the ranks within block is easy in JMP. Just use the Distribution platform with your original response as Y and your blocking variable as a By variable. Then, in the resulting report, hold down the Ctrl key and click on the little red triangle at the top of one of the histograms and choose Save->Ranks. You'll get a new column in your data table with the ranks within levels of your blocking variable.
I hope that helps!
-Jeff
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Friedman test on JMP?
Thank you very much Jeff. However I am not sure I have found what I am looking for: 2-way ANOVA. It seem in the end I receive only one-way ANOVA data in the output file. This is what I did: First, I conducted Distribution analyses and rank transformed data from one of the traits I have studied (Y) accoridng to its randomized block design, i.e. block (By). The data was saved as ranks (six rank/block data compiled into one column of the Jmp data sheet), and next I analyzed the rank transformed trait with Fit Y by X: Y as rank data, and for X I used treatment as a variable. However, in the output file I only see one-way ANOVA data!
Have it been conducted the wrong way? Or is the data to be find somewhewre within the output file?
Thankful for input in this matter!
Best regards,
Jim
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Friedman test on JMP?
For X, you must specify both treatment and block (but not their interaction) to get Friedman's test.
Steve Denham
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Friedman test on JMP?
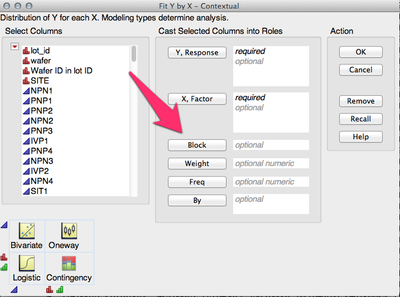
Hi Jim,
You need to specify your Blocking variable in the Block role in the Fit Y by X dialog:
-Jeff
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Friedman test on JMP?
Hej!
Thanks for all the help. I will get into the matter and hoipefully be able to solve it soon. I will be back with some hopefully successfull results.
Sorry for late reply, I am busy with some lab work.
Sincerely,
Jim
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Friedman test on JMP?
Hi again!
Thank you. Unfortunately, I havent sorted it out yet. However, perhaps I need to redo the ranks (distribution analyze to save ranks), since when I run the Fit y by x-analyses a post shows up saying "Blocking ignored because cell counts were unequal." After that I excluded a few replicates that contained some empty cells thinking this could be the reason of unequality, saved it as ranks and ran it in fit y by x, but the unequality still remain and the post shows up again). Could it be that there are some species having 6 replicates and, now some are having 5 replicates? All replicates however are equal in number of treated samples (i.e. control and treated). Is this posing a problem for conducting the Friedman test?
In my Fit y by x-runs I used, in the specification model, the trait values as response Y, and the treatment and block number for factor X and block. I even switched the treatment and block number for those two specifications. But in addition to receiving the previously mentioned post sign it did not display any data or relevance (2-way ANOVA p-values for example).
Also. So far. The ensuing output file doesnt (never) display any two-way ANOVA data, but instead the output file (as always) have the caption of One-way ANOVA. How exactly can you see whether your Friedman test is shown or not on the output file? I dont think there is a caption saying, for example, nonparametric two-way ANOVA in the fit y by x-output?
If anyone have a clue on this please drop a line or two! I am not giving up on this one (yet).
Best regards,
Jim
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Friedman test on JMP?
Give up NOW on Friedman's Test. It only applies when there is an equal number of observations in each block. Think about it--consider two blocks, one with five observations and one with ten. The mean of the first block is 3, of the second 5.5. You automatically have a difference, so the null hypothesis cannot be true, thus any p values are, well, bogus. This means you can't just rank transform your data by block, and get anything meaningful. JMP protects you from doing this, and issues the error message.
So it is time to stop, and reconsider your analysis approach. You say you have non-normal data. That may be, but the assumptions of ANOVA are that the errors are normally distributed. Have you examined them? What does the error distribution look like? Is it amenable to transformation? It is probably time to consult with someone face-to-face to try and answer these questions. Then, an appropriate analysis plan can be devised.
Steve Denham
Recommended Articles
- © 2026 JMP Statistical Discovery LLC. All Rights Reserved.
- Terms of Use
- Privacy Statement
- Contact Us