- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Discussions
Solve problems, and share tips and tricks with other JMP users.- JMP User Community
- :
- Discussions
- :
- Re: Freshly made Definitive Screening Design does not have foldover or center po...
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Freshly made Definitive Screening Design does not have foldover or center points
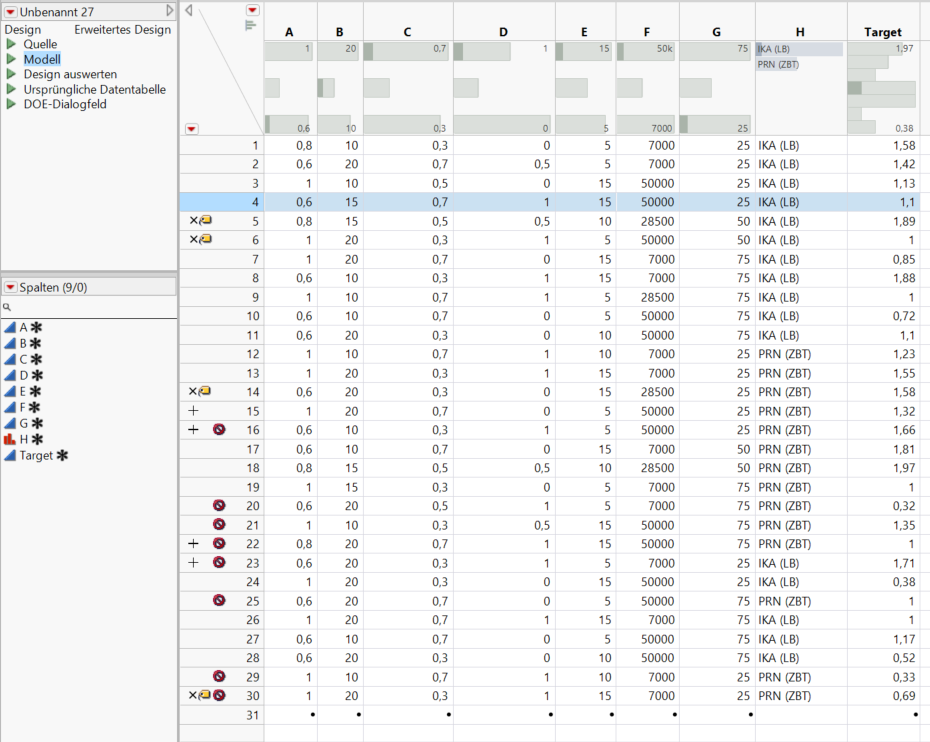
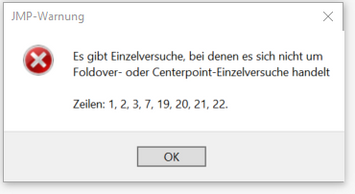
I have designed an DSD with 7 continuous variables and a one categoric variable. 18 runs + 4 extra runs. some of the runs however failed, because of a reactive interaction of the solvent with the high temperature, we decided to skip them, thatfore replicate some other important ones. Now, when the results are there and I am about to analyze by the DSD method - 8 runs for some reason are not center-point or foldover-experiments.
I have made the design from the start, again the same problem.
How I even can identify the center runs? I have not changed anything on the values of the factors.
Where is the mistake? How it is possible to solve that issue?
Greetings
Edward
Accepted Solutions
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Freshly made Definitive Screening Design does not have foldover or center points
Hi @EdwardN ,
A DSD has the unique property that all runs are either:
- centre points (all factors set to the mid-point) or
- foldover pairs
Foldover pairs are 2 runs that are exact mirror images of low and high settings. For example, this is a foldover pair:
- -1, 0, +1, +1
- +1, 0, -1, -1
You should find that in your original design with 22 runs, all your runs are either centre-points or they are part of a foldover pair.
By excluding some runs and repeating others, this will no longer be the case.
The Fit DSD analysis requires the foldover structure. When you try to analyse your modified experiment with Fit DSD JMP tells you that this is not possible because you have runs that are not part of a foldover pair (or a centre point).
You can still analyse your experiment but you will need to use Fit Model.
I hope this helps,
Phil
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Freshly made Definitive Screening Design does not have foldover or center points
Hi @EdwardN ,
A DSD has the unique property that all runs are either:
- centre points (all factors set to the mid-point) or
- foldover pairs
Foldover pairs are 2 runs that are exact mirror images of low and high settings. For example, this is a foldover pair:
- -1, 0, +1, +1
- +1, 0, -1, -1
You should find that in your original design with 22 runs, all your runs are either centre-points or they are part of a foldover pair.
By excluding some runs and repeating others, this will no longer be the case.
The Fit DSD analysis requires the foldover structure. When you try to analyse your modified experiment with Fit DSD JMP tells you that this is not possible because you have runs that are not part of a foldover pair (or a centre point).
You can still analyse your experiment but you will need to use Fit Model.
I hope this helps,
Phil
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Freshly made Definitive Screening Design does not have foldover or center points
Hi @Phil,
ok, I see, that makes sense. So I either can than manually add foldovers to the extra experiments, or have to live with a much smaller set of experiments, because the foldovers have to come in pairs righ? I thought adding data can just make it better - so its basically not possible to add replicate runs manually?
The target is not that complete - I ve added "1" where there was no data, when somehow desperately trying to solve the problem :ll- also the excluded do not really count - we are manufacturing PEMFC- Membrane electrode assemblies by the "indirect" method - that means the electrode is transfered from a teflon sheet to the membrane in a hot pressing step, in some cases this transfer step is not working properly for different reasons meaning the target value is not really a function of most of the variables I would say, like loading of the platinum-catalyst etc. In this case the fit gets really bad, thatfor excluded. As you ve proposed I fit the data with fit model. Actually even with lot of experiments missing I got a more or less decent fit, but still theres plenty room for improvement.
Is there a possibility to highlight the foldover pairs? Or check the design for missing pairs somehow?
Thank you very much for the help!
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Freshly made Definitive Screening Design does not have foldover or center points
Hi @EdwardN ,
I would not recommend trying to add the foldover pair of your additional runs. The foldover structure of a DSD is useful and it enables Fit DSD, which is also useful. But trying to force your problem into a foldover design does not make sense.
Instead I would recommend that if you are going to add more runs, you use the Augment Design platform. You can specify the factor ranges that you think are going to work. And JMP will give you an optimal set of runs to estimate the model that you specify.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Freshly made Definitive Screening Design does not have foldover or center points
Hi Phil,
ok, that makes pretty much sense. However theres another problem appearing: in the next experimentation round we want to reduce the amount of factors, but we also noticed another factor we kept constant until now, but now want to include it. In the Augment design I can just choose from already existing splits. So can I just draw a new split, insert constant values there, and then add it to the Augment Design? Will JMP undarstand that? Or would you recommend to better start a completely new screening round?
Greetings
Edward
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Freshly made Definitive Screening Design does not have foldover or center points
A very good questions and it has been asked and answered before.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Freshly made Definitive Screening Design does not have foldover or center points
This is a separate issue but I wonder if you really needed to exclude the runs where you had a reactive interaction with the solvent. It looks like you have response data ("Target") for those runs, so you can still include them in a model.
We design experiments to learn. And often we can learn just as much from the bad results. As long as you have reliable response data for these runs you should not exclude them.
Recommended Articles
- © 2026 JMP Statistical Discovery LLC. All Rights Reserved.
- Terms of Use
- Privacy Statement
- Contact Us