- We’re retiring the File Exchange at the end of this year. The JMP Marketplace is now your destination for add-ins and extensions.
- JMP 19 is here! Learn more about the new features.
- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Discussions
Solve problems, and share tips and tricks with other JMP users.- JMP User Community
- :
- Discussions
- :
- DoE How to treat replicate measurements
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
DoE How to treat replicate measurements
(I needed to extend the question, the update is below the images)
Hey everyone,
I am looking for some advice. I have performed a definitive screening design (JMP 11) with 3 factors, 1 response (9 experiments) three times. I augmented the design and entered the new data. The problem is, that some combinations produce very varying results. I have repeated some of these experiments additional three times and the variation seems to be "inherent".
My question is how to treat these replicate runs. When I augment the design to have 2 replicate runs (3 runs in total), my regression looks pretty bad.
However, when I average my trial runs first and then perform the regression, I get much better results.
But when I do the averaging first, I think I disregard the information about the standard deviation/variance, which I would like to include (and maybe penalize, because we would like to work in a "stable" region).
Is there any way to include this into the DoE? Maybe use the SD as a response and minimize it?
Edit:
I've thought about my problem and would like to extend it a "little" bit. I thought that I could use the SD as weight for my data, in order to take into account that some conditions used for the experiments lead to very varying results and others are more reproducable. So I read about weighted least square regression and now I am very confused because choosing the weight appears to be non-trivial. Thus my question extends to: If I use a weight, which one would be appropriate. Currently I have thought about and tried the following approaches:
- Use the SD (from what I have seen, it should used as 1/SD or 1/SD^2). This sounds very reasonable to me but on the other hand I have read that one needs lots of replicates to have a proper estimate of the SD (in the range of dozens), otherwise this method is not accurate enough.
- Use one of my responses as a weight. I am also measuring the so called polydispersity index, which gives me some information about the homogenity/heterogenity of my samples. The results looks quite good, but I am not sure if it is allowed to use a response a weight (I think I would introduce some kind of bias?)
- Use the residuals. I have found this in a lecture handout. The idea is to make a non-weighted fit first and the use 1/(residuals)^2 as the weight. But againg, I am worried to "push" the resulting fit into a wrong direction.
Thanks a lot!
Nachricht geändert durch Roland Goers
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: DoE How to treat replicate measurements
Regarding the "Rep" factor:
In which way do I need to integrate it into my modeling? I usually choose my three factors, click the RSM macro in JMP and proceed. Do I need to include the "rep" factor into the RSM macro or leave him out (Thus no interactions with other factors)?
Futhermore, if the "rep" factor is considered significant and I have to include it into the model, it also appears in the profilers. However, this is kind of confusing for me, because on the one hand I understand that this factor is significant and thus my results are depending on the batch. This makes perfectly sense, I have a biological component in there, they are never the same. On the other hand, if I now optimize my results, the batch is also included and for example batch #1 does not exist anymore....
Do I miss something? Is there a way to include the batch variance without including a batch variable?
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: DoE How to treat replicate measurements
After you create your RSM with your design variables, add the Rep variable. At this point it is considered a Fixed effect, and will appear in the model equation. Go ahead and do the regression this way first. You can check to see if it is significant, and how big an effect it is. If it is significant, transform the Rep variable to a 'Random Effect' (click the Attributes red triangle), and do the regression again. This time it will not appear in the model equation, rather, it is considered a random effect. "A random effect is a factor whose levels are considered a random sample from some population. Often,...variance components). However, there are also situations where you want to predict the response for a given level of the random effect. Technically, a random effect is considered to have a normal distribution with mean zero and nonzero variance."
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: DoE How to treat replicate measurements
Thank you very much for your answer!
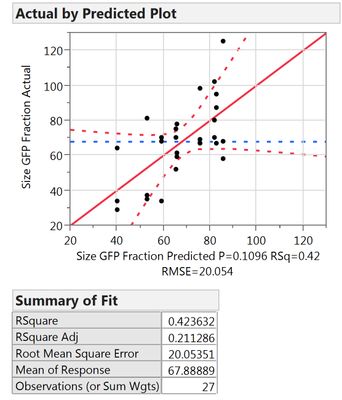
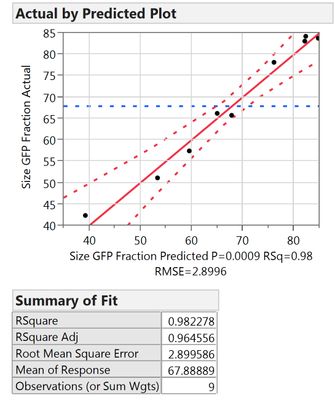
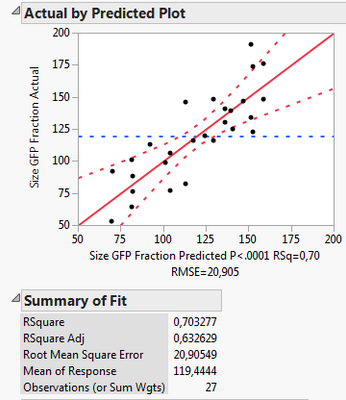
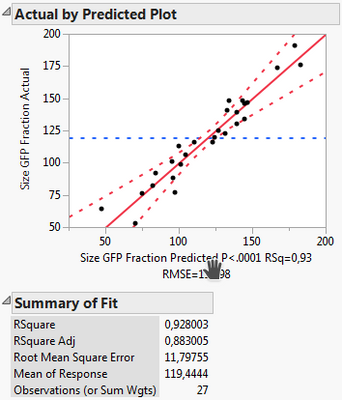
Out of curiosity, why is the "rep" factor not included into the RSM (or not allowed to be)? I just compared the two results
Following your guidelines:
Including it into the RSM:
What I can see, is that the first model includes less factors, namely X1, X2, X3, X3*X3 and Xrep, whereas the second model uses X1*Xrep, X2*Xrep and X3*Xrep additionally. Thus it has more terms and might overfit. However, all criteria like Rsq adj., BIC and AICc are also better for the 2nd model.
The results of the optimization are very similar.
Using simply common sense, I would imagine that during each replicate run, the input factors varied which these terms would take into account.
Or am I totally wrong and this simply forbidden?
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: DoE How to treat replicate measurements
I like to look at the higher XRep terms as a diagnostic, for example to help figure out what may be happening and maybe to reduce the effects in the future. Based on the factors you listed, it looks like there may have been something that occured over time. But I would not include anything except the XRep as a random effect in the final model. As you know, XRep is not something that can be asigned in future simulations.
hope this helps,
mark
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: DoE How to treat replicate measurements
I would first add a new nominal variable called 'Rep' or 'Run' and tabulate which set of replicates were run together, for example 1,2,3 etc. Now include this your model analyses. Start by adding this new variable as a random or fixed effect. This will assign some of the rep to rep variation, but does little to diagnose it. You can add more complex effects such as rep-to-rep interactions to explore what may be causing the variation.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: DoE How to treat replicate measurements
Thank you all very much for your help and detailed explanation! Especially cipollone.mg/Mark!
- « Previous
-
- 1
- 2
- Next »
Recommended Articles
- © 2025 JMP Statistical Discovery LLC. All Rights Reserved.
- Terms of Use
- Privacy Statement
- Contact Us