先日、日本のお客様向けに「JMPによるスペクトルデータ/クロマトグラムデータの多変量解析」というタイトルのセミナーを実施しました。
このセミナーは、タイトルの通りスペクトルやクロマトグラムデータを扱うニッチな内容だったため、参加者は少ないだろうと勝手に思っていました。しかし、想像を大きく覆すほどの多くの方にご参加いただきました。有難い限りです!
セミナーの中では、主に以下のラマンスペクトルのデータを扱いました。
分析の目的
ウコン中に含まれる鉛の含有量をラマン分光法(散乱モード)を用いて定量するためのモデルを開発する
スペクトルデータの概要
- 鉛濃度が異なるように調製された42種類のウコンを含む試料を準備
- 鉛の濃度によって、A~Fの6つのグループ(各グループ7つの試料)に分類
以下の図は、42種類の試料のラマンスペクトルデータを「グラフビルダー」を使って視覚化したものです。凡例にある通り、グループごとに色分けをしています。
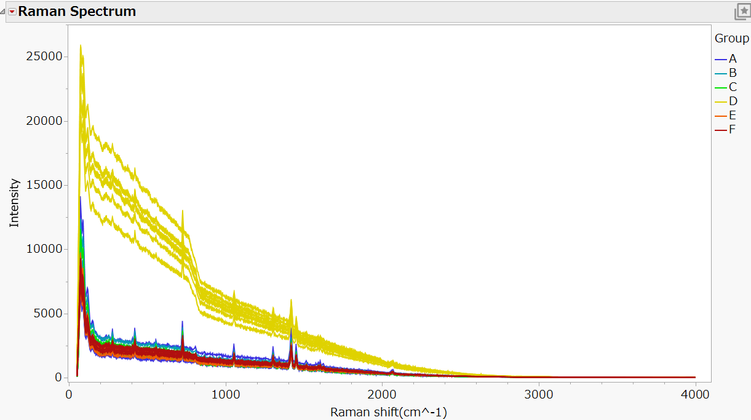
データの出典:Cruz, Jordi et al. (2020), “Data for: Quantitative models for detecting the presence of lead in Curcumin using Raman spectroscopy”, Mendeley Data
関数主成分分析を実施してスペクトルを分類
セミナーでは、このデータを用いてPCAやPLS回帰を実行してモデル化を行い、さらに応用例として関数主成分分析(FPCA)を用いたスペクトルの分類方法についても紹介しました。
関数主成分分析は、JMP Proの「関数データエクスプローラ」プラットフォームで実施できます。そこで、これらのスペクトルデータにウェーブレットモデルをあてはめ、関数主成分分析による「スコアプロット」を考察しました。このスコアプロットにより、試料のスペクトルの形状に基づく類似性を視覚的に確認できます。
近い位置にプロットされた試料は形状が似ており、遠くにプロットされているものは形状が異なります。そのため、スコアプロットでグループごとにまとまってプロットされれば、ウコンの濃度で試料を分類できていることになります。
下図のスコアプロットを見ると、黄色のプロット点(グループD)は成分1の右側に位置し、他のグループは左側に位置しています。(スペクトルの形状を視覚的に把握するため、以降のスコアプロットでは、3つの試料A7,D4,D7についてはポッポアップで表示できるスペクトルの形状を示しています。)

このことから、グループDは他のグループ(A,B,C,E,F)と明確に区別できましたが、グループA,B,C,E,Fは完全には分けられていないことがわかります。
スペクトルデータを扱っている方はピンと来たかもしれませんが、上記の例はスペクトルデータの前処理をせず、測定されたローデータで分類を試みた結果なのです。
スペクトルデータの前処理
スペクトルデータの分析において前処理(プリプロセッシング)が重要だと言われています。前処理を行うことでノイズの低減や不要な変動の除去、特徴を強調させるなどが可能となり、分析精度の向上が期待できます。
JMP Proの「関数データエクスプローラ」では、スペクトルデータで良く用いられる前処理の機能が搭載されています。

それぞれのコマンドの詳細は以下の通りです。
| 手法 |
内容 |
| SNV |
サンプルごとに標準化処理(平均を0、標準偏差を1に)する |
| MSC |
乗算的散乱補正、サンプルごとに単回帰をあてはめ補正する |
| Savitzky-Golay |
SGフィルタを適用し、適用したものに1次微分、2次微分を行う |
| ベースライン補正 |
個々の関数からベースライン関数を差し引いて補正する |
前処理後の分類結果
まず、ローデータにSavitzky-Golayフィルタ(SGフィルタ)を適用し、1次微分を行う前処理を実施しました。
SGフィルタとは、いくつかの点のまとまりに多項式をあてはめて平滑化処理をする方法です。多項式なので微分処理することができ、1次微分を行うことによりピークの傾斜(変化率)が強調されます。
この前処理をしたデータにウェーブレットモデルをあてはめ、関数主成分分析におけるスコアプロットを確認しました。D以外のグループの試料は先ほどに比べばらついてプロットされていますが、まだグループを分類できている状態ではありません。

さらに、SGフィルタ適用したデータにSNVの処理を実施し、同様の分析でスコアプロットを確認しました。
すると、グループAが他のグループからはっきりと分離されていることが確認できました。

最終的に、関数主成分1, 2に加え、成分3を含めた三次元でのプロットを行い、グループDおよびグループAが他のグループから分離できていることが確認できました。一方で、グループBとCはほぼ重なっており、完全には区別できませんでしたが、グループEとFは成分3まで見ることである程度の分類が可能でした。

グループDとグループAは、他のグループから分かれており、グループB,Cはほぼ重なっているので分けられていない、EとFは成分3(Y FPC3)までみると、完全でないにせよある程度分けられていることが分かります。
この事例を通じて、スペクトルデータの前処理がいかに重要であるかを再確認しました。前処理により、データの質を向上させ、より正確な分類結果を得ることが可能になりました。
参考:関数データエクスプローラについて
関数データエクスプローラの概要 ~関数主成分分析とさまざまな利用事例~ (日本語) - JMP User Community
by 増川 直裕(JMP Japan)
Naohiro Masukawa - JMP User Community
You must be a registered user to add a comment. If you've already registered, sign in. Otherwise, register and sign in.