- Subscribe to RSS Feed
- Mark as New
- Mark as Read
- Bookmark
- Subscribe
- Printer Friendly Page
- Report Inappropriate Content
JMP Add-Ins
Download and share JMP add-ins- JMP User Community
- :
- File Exchange
- :
- JMP Add-Ins
- :
- Data visualization with t-SNE and UMAP
Description
Recently, non-linear dimension-reduction and visualization algorithms, most notably t-Distributed Stochastic Neighbor Embedding (t-SNE) and uniform manifold approximation and projection (UMAP), have been widely applied to various research areas such as image processing, text mining, and genomics. This Add-in provides access to both t-SNE and UMAP R packages. It offers a user-friendly interface enabling data table navigation, data quality control, sparsity handling, intuitive parameterization, and interactive results interpretation.
Usage Example
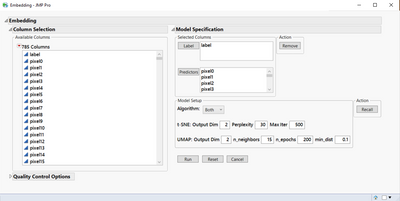
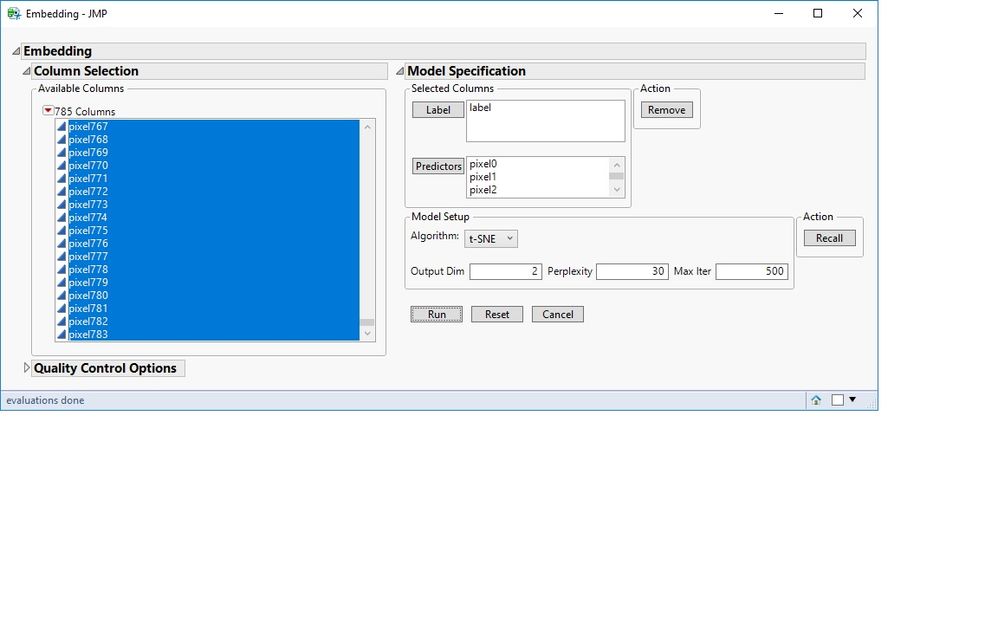
Here is a screenshot of the interface with MNIST data loaded. Under Model Specifications, I selected the label column as Label and all the pixels as predictors. I chose both t-SNE and UMAP as the algorithms.
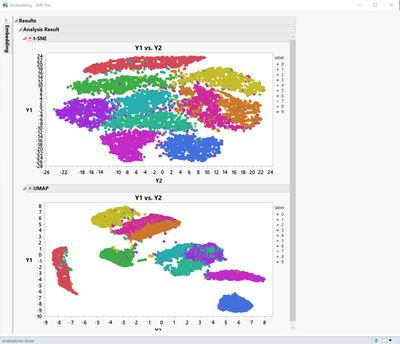
Another screenshot of the results of both t-SNE (top) and UMAP (bottom).
This add-in also supports some basic quality control options, including missing value checking, distribution, and sparsity calculation. You can find these options under Quality Control Options on the interface.
Updates: JMP R interface on Mac has versioning issues. Please downgrade your R to version <=3.3.3, and use t-sne only if you are a Mac user.
Changelog:
v.2 <9/22/2020> Added tsne and umap results to the original table and reorganized output for better exploration experience.
v.1.2 <3/14/2019> Fixed an issue in Rtsne package where a large number of columns causing stack overflow problem.
v.1.1 <3/8/2019> Fixed a bug that could potentially produce “issues found in R, memory exhausted?” error message. Added a submenu for the MNIST example dataset.
v.1.0 <2/26/2019> Initial version.
Hi! MJ,
Can I understand that, in above photos, each mass/group of points can be treated as the same attribute? or different color points should be treated as different attribute?
The colors indicate true labels for this dataset, so data points with the same color should have similar attribute. The clusters were estimated by t-SNE and UMAP, you should see some color mismatches.
Hi MJ,
I am getting an error message when I try this addin. It looks like it is not recognizing my installation of R. Would you be able to help figure out how to get your addin to recognize my R installation if that is indeed the issue?
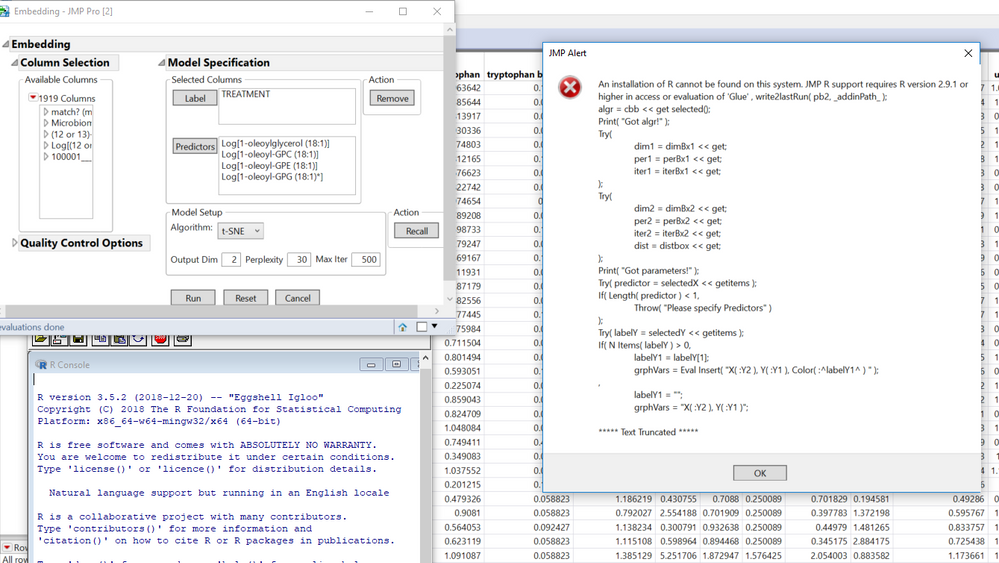
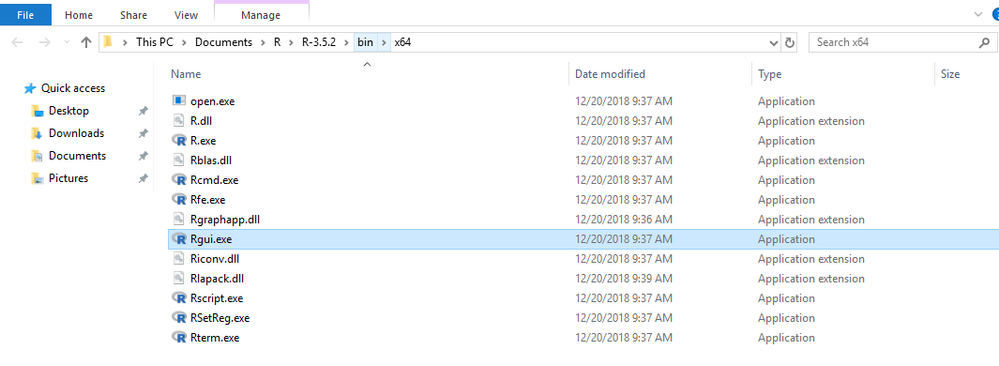
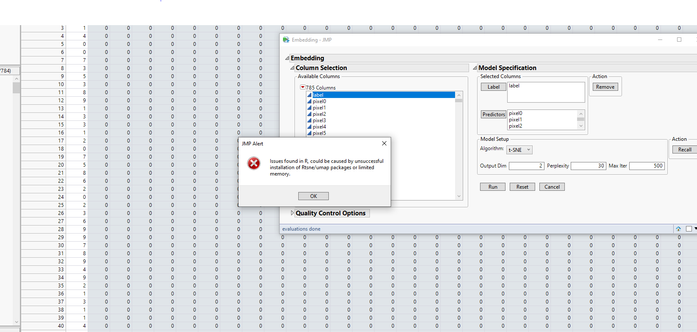
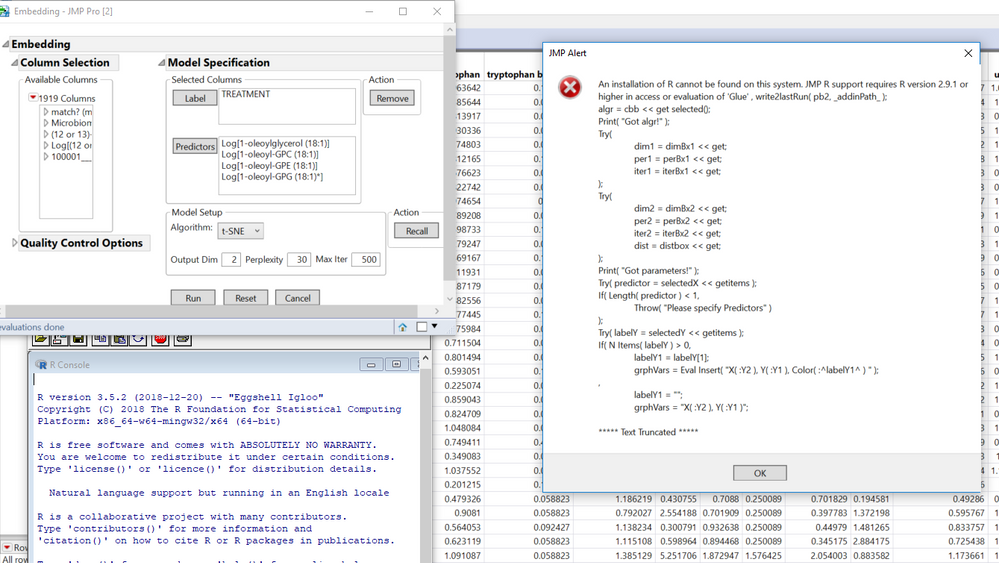
I am including a screenshot showing that R is open but not being recognized, and a screenshot of my R install location (not program files) and also copying some of the error message below.
This looks like a very exciting tool and I am hoping to use it. I've previously found t-sne in R to be useful and it would be of great value to be able to do this right in JMP. Any help you might provide is greatly appreciated.
Thanks!
Screenshots of error and open R instance, then screenshot of R install location
Error code
"
An installation of R cannot be found on this system. JMP R support requires R version 2.9.1 or higher in access or evaluation of 'Glue' , write2lastRun( pb2, _addinPath_ );
algr = cbb << get selected();
Print( "Got algr!" );
Try(
dim1 = dimBx1 << get;
per1 = perBx1 << get;
iter1 = iterBx1 << get;
);
Try(
dim2 = dimBx2 << get;
per2 = perBx2 << get;
iter2 = iterBx2 << get;
dist = distbox << get;
);
Print( "Got parameters!" );
Try( predictor = selectedX << getitems );
If( Length( predictor ) < 1,
Throw( "Please specify Predictors" )
);
Try( labelY = selectedY << getitems );
If( N Items( labelY ) > 0,
labelY1 = labelY[1];
grphVars = Eval Insert( "X( :Y2 ), Y( :Y1 ), Color( :^labelY1^ ) " );
,
labelY1 = "";
grphVars = "X( :Y2 ), Y( :Y1 )";
***** Text Truncated *****"
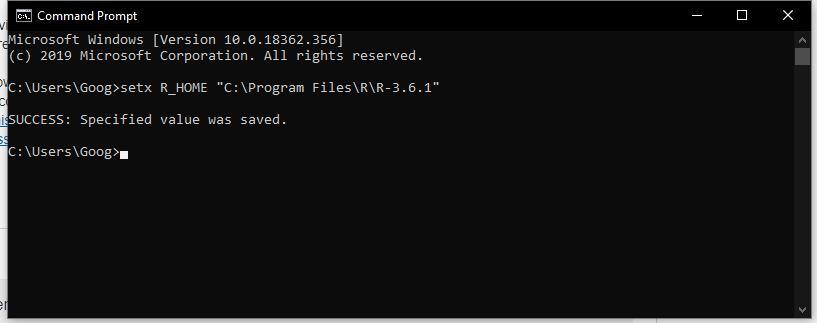
Your problem was likely caused by multiple installations of R on your machine and JMP couldn't decide which one to use. Please try to set R_HOME as an environmental variable as following and try this add-in again. Open Window CMD console and type: setx R_HOME "This PC\Documents\R\R-3.5.2". And make sure Rtsne and umap packages are installed to this version of R. Please let me know if this solves your problem.
Also, we have a few threads on the community talking about this issue that you can check out.
https://community.jmp.com/t5/Administration-Discussions/Help-JMP-find-R-installation/td-p/6357
https://community.jmp.com/t5/Discussions/Setting-path-to-R-location/td-p/59764
Hello @MJ,
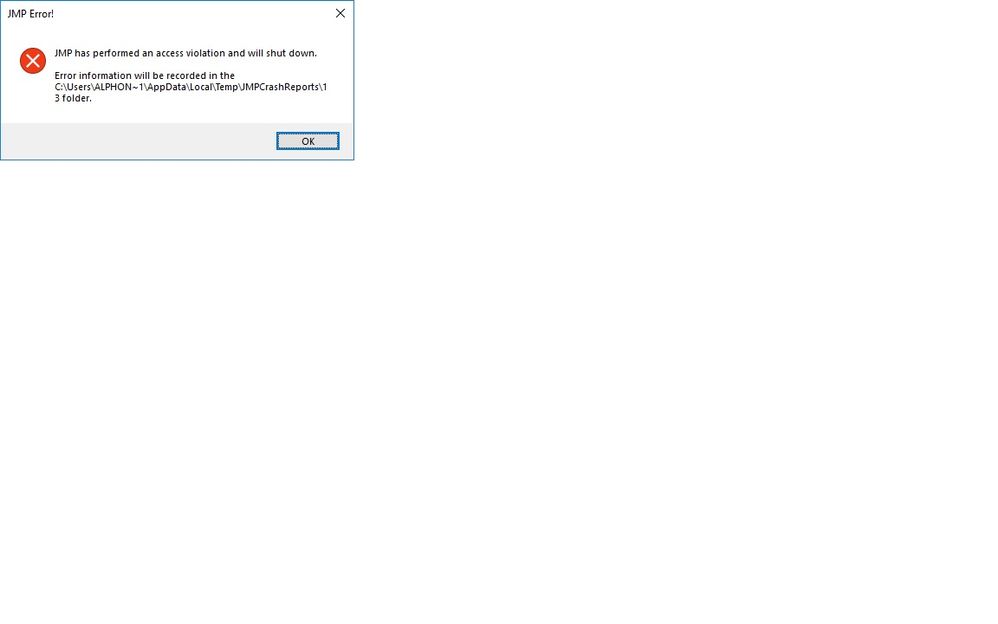
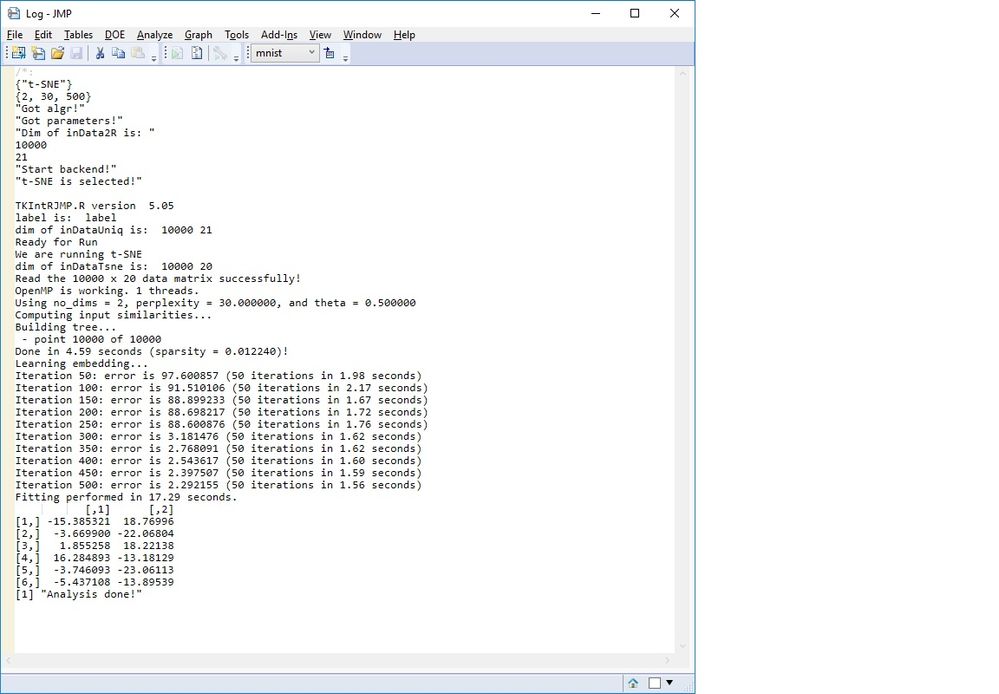
I tried running your sample data, but had a access violation crash. In the log file, it looks like the R-side ran successfully. Any ideas how I can address the issue?
Thanks,
Al
Hi @ngalphie, this looks like a problem with the older version of JMP.
1. I noticed that you are using JMP 13, it might be helpful if you can update it to the latest version (JMP 14.3) and try this add-in again.
2. Send the crash report saved at C:\Users\username\AppData\Local\Temp\JmpCrashReports\13 to me to tech support (https://support.sas.com/ctx/supportform/createForm?ctry=us_JMP) or me directly (Meijian.Guan@jmp.com), we can dig into it.
3. There are a few posts on the community also talking about this issue. You can take a look to see if there are anything they mentioning could help.
https://community.jmp.com/t5/Discussions/JMP-Access-Violation-Likely-Causes/m-p/5648#M5647
Thank you very much for providing an interface for JMP.
I wonder if these addins can include the R/Python executables so we can run them directly.
@FN Hey there, thank you for your suggestions. It would be a great option to include R/Python executables but due to our policies and legal concerns, I did not include them. Please let me know if you have any problems regarding R installations or versions when using this add-in.
Thank you. To be honest, I am not sure what is the best way to install R. I am used to manage Python installations with conda/anaconda, which also includes R now.
This is the path where I have R installed.
(base) C:\Users\john_doe>where r
C:\Users\john_doe\AppData\Local\Continuum\miniconda3\Scripts\R.exe
I guess I need to install these packages
https://anaconda.org/conda-forge/r-tsne
https://anaconda.org/conda-forge/r-umap
To make JMP able to find my R installation, I exectue this (or change the PATH manually):
setx R_HOME "C:\Users\john_doe\AppData\Local\Continuum\miniconda3\Scripts\"
If there is a detailed guide on how to do this better, please let me know.
I managed to run umap but not via Anaconda/conda.
I think I am installing the wrong package for tsne. Can you provide the URL in cran?
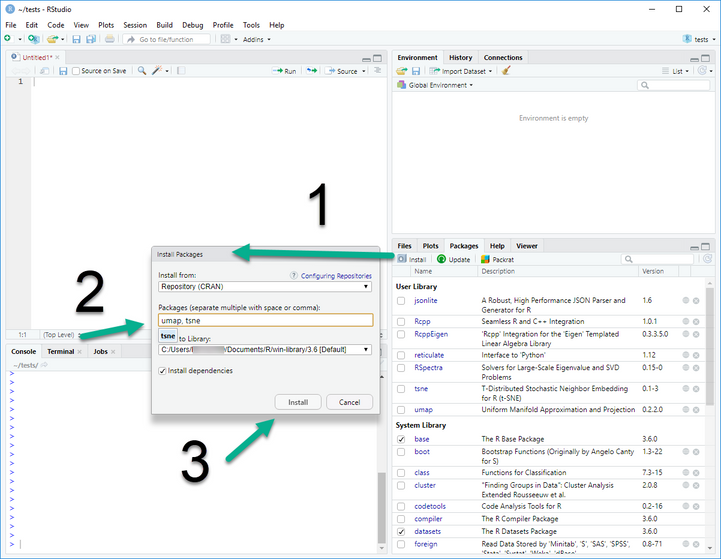
Here is the step by step.
Intall R from https://cran.r-project.org/
Install Rstudio commnutiy https://www.rstudio.com/products/rstudio/download/#download
Use Rstudio to install tsne and umap.
Hello, @FN, it looks like you installed a different version of t-sne package. Could you please try to install Rtsne through R studio instead? The github version for this package is here: https://github.com/jkrijthe/Rtsne. Also please make sure your R_HOME path is pointing to the right R version with UMAP and Rtsne installed. Let me know if you have further questions.
Dear @MJ
thanks for this nice add-in. I used it on a Mac tSNE with R Version 3.3.3 worked fine. UMAP did not.
Now I checked it on Windows using the latest R Version 3.6.1 with RStudio. I installed the packages for umap and Rtsne unfortunately neither umap nor Rtsne worked. I use JMP 14.3.
Here is what log says when I try the mnist data after PCA (using 2 PCs as predictors):
{"UMAP"}
{2, 3, 200, 0.1}
"Got algr!"
"Got parameters!"
"Dim of inData2R is: "
10000
3
"Start backend!"
"UMAP is selected!"
TKIntRJMP.R version 14.0
label is: label
dim of inDataUniq is: 10000 3
Ready for Run
We are running UMAP
An exception of type c0000005 occurred at address 6c910ef2 while processing the submitted R statements. This address is at offset 10ef2 into module "C:\Program Files\R\R-3.6.1\bin\i386\R.dll"
An exception of type c0000005 occurred at address 6c910ef2 while processing the submitted R statements. This address is at offset 10ef2 into module "C:\Program Files\R\R-3.6.1\bin\i386\R.dll"
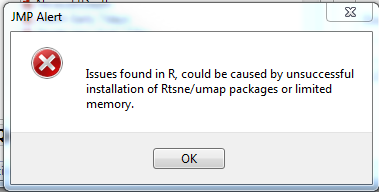
Issues found in R, could be caused by unsuccessful installation of Rtsne/umap packages or limited memory.
{"t-SNE"}
{2, 5, 500}
"Got algr!"
"Got parameters!"
"Dim of inData2R is: "
10000
3
"Start backend!"
"t-SNE is selected!"
TKIntRJMP.R version 14.0
label is: label
dim of inDataUniq is: 10000 3
Ready for Run
We are running t-SNE
dim of inDataTsne is: 10000 2
Read the 10000 x 2 data matrix successfully!
OpenMP is working. 1 threads.
Using no_dims = 2, perplexity = 5.000000, and theta = 0.500000
Computing input similarities...
Building tree...
- point 10000 of 10000
Done in 0.45 seconds (sparsity = 0.001695)!
Learning embedding...
Iteration 50: error is 120.349392 (50 iterations in 2.08 seconds)
Iteration 100: error is 103.025177 (50 iterations in 1.94 seconds)
Iteration 150: error is 94.995130 (50 iterations in 1.70 seconds)
Iteration 200: error is 90.828601 (50 iterations in 1.75 seconds)
Iteration 250: error is 87.964178 (50 iterations in 1.81 seconds)
Iteration 300: error is 4.069311 (50 iterations in 1.82 seconds)
Iteration 350: error is 3.474366 (50 iterations in 1.83 seconds)
Iteration 400: error is 3.044447 (50 iterations in 1.84 seconds)
Iteration 450: error is 2.719790 (50 iterations in 1.83 seconds)
Iteration 500: error is 2.466420 (50 iterations in 1.84 seconds)
Fitting performed in 18.44 seconds.
[,1] [,2]
[1,] -18.4173747 30.133875
[2,] -23.5082997 -17.985255
[3,] 0.7376548 -4.148267
[4,] -5.6881024 6.385878
[5,] 8.8457460 22.245393
[6,] -10.8212058 -17.013940
[1] "Analysis done!"
An exception of type c0000005 occurred at address 6c910ef2 while processing the submitted R statements. This address is at offset 10ef2 into module "C:\Program Files\R\R-3.6.1\bin\i386\R.dll"
Issues found in R, could be caused by unsuccessful installation of Rtsne/umap packages or limited memory.
Would be great if you would have an idea what I should try. Thanks in advance and best regards Patrick
Hi Meijian,
Good afternoon. I tried to use t-SNE and UMAP on the sample data. I use the latest version of R Studio and JMP 13. But I got the following error-
Please me know how can I fix it. Thanks for your help.
PS. I won't be able to upgrade to JMP 14 at this moment.
Sincerely,
Arif
dear Sir
the addins does not run
Dear @Raaed, I believe I have responded above regarding this issue. It's likely because JMP R Interface had trouble finding your R installation.
Please try to set R_HOME as an environmental variable as following and try this add-in again. Open Window CMD console and type: setx R_HOME "Path to R". And make sure Rtsne and umap packages are installed to this version of R. Please let me know if this solves your problem.
If you are using Mac, you need to downgrade your R to version 3.3.3 and only use T-SNE.
Also, we have a few threads on the community talking about this issue that you can check out.
https://community.jmp.com/t5/Administration-Discussions/Help-JMP-find-R-installation/td-p/6357
https://community.jmp.com/t5/Discussions/Setting-path-to-R-location/td-p/59764
Hope that helps!
Dear sir:
after install tsne, Rtsne & umap packages, and type the below script on CMD, the add-ins are run✌
thank you very much
Dear @MJ,
When attempting to run the program on the test data set, I got an error saying "Issues found in R, could be caused by unsuccessful installation of Rtsne/umap packages or limited memory."
I'm pretty sure I downloaded everything I need (the packages and R, too), so if you could clue me into why this isn't working, that would be much appreciated
Regards,
Dhruv Bhattaram
Hi @DBhattaram, it's possible that JMP didn't find the right R version, or the R versioning issues with JMP R Interface. Could you please open the log file (CTRL+Shift+L) when you see the error message and send the detailed log info to me at Meijian.Guan@jmp.com? I'd be happy to take a look.
MJ
Rtsne nicely worked.
Best wishes
Pat
Hi, @MJ
I sent the log of all my failed attempts at getting it to run the program. Hopefully, that will be of some use
Regards,
Dhruv
Thank you @DBhattaram, it looks like you didn't have Rtsne and UMAP package installed to the R version JMP is talking to. If you have multiple versions of R, make sure you set up R_Home as environmental variable as following: open Window CMD console and type: setx R_HOME "your Path to R". And make sure Rtsne and umap packages are installed to this version of R. Let me know if it solves your problem.
Hello, I have the same error saying "Issues found in R, could be caused by unsuccessful installation of Rtsne/umap packages or limited memory." I installed UMAP and TSNE from Rstudio. I use R x64 3.5.2 under Windows
Hello @Lu, sorry for lacking details in the error message. Could you double-check if you installed Rtsne instead of TSNE package in R? Let me know if that fix your problem.
I did install Rtsne as suggested but get the following error message now;
@Lu Your problem was likely caused by multiple installations of R on your machine and JMP couldn't decide which one to use. Please try to set R_HOME as an environmental variable as following and try this add-in again. Open Window CMD console and type: setx R_HOME "This PC\Documents\R\R-3.5.2". And make sure Rtsne and UMAP packages are installed to this version of R. Please let me know if this solves your problem.
I am a Windoxs 7 user and not an ICT expert. So I do not find how to get into the "CMD console", sorry :-(. I removed R en reinsatlled it in the same map as JMPpro. Still getting the same error message as above. Not a good idea?
@Lu Sorry for the trouble. On Windows 7, I think you can go to the start menu, type cmd or command in the search box, and the press enter. Or you can just go to Start Menu and go to All Programs. Then, open Accessories and click the Command Prompt shortcut.
Still receiving the following error when executing the Embezdding Add-in.
Add-in still not working in JMP-pro after changing the CMD. In R, I used the Tab "Packages"- "Install Packages" to install umap and Rtsn package. Any other suggestion?
Any other suggestion
So, is t-SNE/UMAP going to be added to a future version of JMP? It is a very powerful abnormal event detection technique with wide applicability across industry. I just read this paper. t-SNE performs very well on the Tennessee Eastman Process dataset:
A new unsupervised data mining method based on the stacked autoencoder for chemical process fault di...
Hi @markschahl, I know John Sall was working on a JMP version of t-SNE but not sure when we are going to release it. I agree that it's a powerful method and implementing it in JMP would be the optimal way as calling out to R has many potential issues. You can request it as a new feature through our technical or product management teams.
@MJ & @markschahl , another thought is to add it to the JMP Wishlist on the community:
https://community.jmp.com/t5/JMP-Wish-List/idb-p/jmp-wish-list
This will help with getting some numbers around the demand and let others voice their support for the capability (I agree it would be great to have in JMP, too!)
M
I have been using the embedding addin in JMP 14+ and JMP15. This addin create tsne and umap images and pca data with an R interface. It is an excellent program and easy to use as it is so quick. The R interface instructions worked well.
I have used it on many databases. However, on a recent smaller database of 74 rows it always throws an error message (not enough memory or incorrect installation). I reinstalled it again but the same problem. Working through data I determined that this addin requires at least 90 rows. This is not a problem with sklearn. I would prefer to use this addin instead of sklearn as this addin is much faster for multiple analyses and readily permits exporting the primary x and y pc data to another jmp file for the manual creation of new classes.
Is there a fix for this?
Love JMP. Thanks, Neal
Hi @rexneal, I'm glad that you found this add-in useful. I will look into this issue and get back to you as soon as I can.
Thanks,
MJ
Hi @rexneal, I did some exploration, it was likely because of the perplexity level you set was too high for a dataset with less than 90 rows. I would recommend reducing it to 5~10 in this case. I will soon post an updated version of this addin allowing better comparison between the two methods.
- Mark as Read
- Mark as New
- Bookmark
- Get Direct Link
- Report Inappropriate Content
Hi,
I get the following error:
The package has been running on different JMP datasets. What specifically triggered this?
Hi @MikeDereviankin, I just published a new version of this Add-in. Could you please try this new version and let me know if it works for you? It seems like there is something wrong with your R installation.
- Mark as Read
- Mark as New
- Bookmark
- Get Direct Link
- Report Inappropriate Content
@MJ
Just installed the add in and now JMP can't find my R ... I've reset my environmetal variables, changed the path in CMD, reinstalled R and nothing.
I get the error saying R is not installed. This doesn't seem as an effective add-in with all these problems that are avoided if T-SNE is just run in R. The GUI is nice, but this seems to be a headache based on all the comments.
In case it helps, this text is from the "How JMP Finds R" section of the JMP 15 scripting guide . From my experience setting the environment variable from within JMP is the safest method.
JMP delays loading R until a JSL-based script requires access to it. When JMP needs to load R,
it follows the standard steps for finding R on a Windows computer:
1. Look up the environment variable R_HOME.
If the variable exists, load R from the specified directory.
2. If the environment variable R_HOME does not exist, look up the InstallPath value in the
Windows registry under the following key:
HKEY_LOCAL_MACHINE\SOFTWARE\R-core\R
If the InstallPath value exists, load R from the specified directory.
3. If the InstallPath value does not exist, an error message states that R could not be found.
Thank you @ih for sharing your personal experience.
@MikeDereviankin My apologies for the R integration issues, usually having single R installation, restart R, JMP, or even your machine would help in this scenario. After you going through these troubles, you might appreciate the convenience and the interactivity of this add-in.
Hi MJ,
Thanks for the Add-in.
Does the predictors have to be integers?
Best,
Ed
Hi @Ed1, no, the predictors can be any type of numeric values. It may not work with characters though.
- Mark as Read
- Mark as New
- Bookmark
- Get Direct Link
- Report Inappropriate Content
@MJ
What does it mean when t-SNE returns "cannot get R output". I've made sure to set my CMD and I even ran JMP as the administrator. Where else could there be a security problem?
- Mark as Read
- Mark as New
- Bookmark
- Get Direct Link
- Report Inappropriate Content
@MJ below is the log:
{"t-SNE"}
{4, 30, 500}
"Got algr!"
"Got parameters!"
"Start inDataPrep"
{"PCB-1_imputed, Pct of Total", "PCB-2_imputed, Pct of Total", "PCB-3_imputed, Pct of Total", "PCB-4_10_imputed, Pct of Total", "PCB-5_8_imputed, Pct of Total", "PCB-6_imputed, Pct of Total", "PCB-7_9_imputed, Pct of Total", "PCB-11_imputed, Pct of Total", "PCB-12_13_imputed, Pct of Total", "PCB-15_imputed, Pct of Total"}
"Finish Sparsity"
"Sparsity is: "
0.00300530234872998
"Label is: "
{"Type"}
"Dim of inData2R is: "
439
120
"Finished data preparation."
"Start backend!"
"Label is: "
{"Type"}
"Within talk2R function."
"label is:"
{"Type"}
"t-SNE is selected!"
TKIntRJMP.R version 14.0
"Sending label to R: "
{"Type"}
"Sending label to R"
data4R=data.frame(data4R)
#label=unlist(label)
#cat("label is: ",label,"\n")
#print(length(label))
#cat("\n")
#remove duplicated observations from both datasets
#inDataUniq=data4R[!duplicated(data4R[,!names(data4R) %in% label]),] #allow excluding multiple labels
#head(data4R)
#cat("dim of inDataUniq is: ",dim(inDataUniq),"\n")
cat("Ready for Run","\n")
if("t-SNE"=="t-SNE"){
cat("We are running t-SNE","\n")
library("Rtsne")
inDataTsne=data.matrix(data4R[,!names(data4R) %in% label])
cat("dim of inDataTsne is: ",dim(inDataTsne),"\n")
tsne <- Rtsne(inDataTsne, dims = 4, perplexity=30, verbose=TRUE, max_iter = 500,pca=F)
outputY=tsne$Y
head(outputY)
}else if("t-SNE"=="UMAP"){
cat("We are running UMAP","\n")
library(umap)
outUmap=umap(data4R[,!names(data4R) %in% label], #method="umap-learn",
n_neighbors=15, n_components=2, n_epochs=200,min_dist=0.1)
outputY=outUmap$layout
head(outputY)
}else if("t-SNE"=="Both"){
cat("We are running Both","\n")
library("Rtsne")
library(umap)
cat("\n","Start t-sne","\n")
inDataTsne=data.matrix(data4R[,!names(data4R) %in% label])
tsne <- Rtsne(inDataTsne, dims = 4, perplexity=30, verbose=TRUE, max_iter = 500,pca=F)
outputY1=tsne$Y
cat("Start umap","\n")
outUmap=umap(data4R[,!names(data4R) %in% label], #method="umap-learn",
n_neighbors=15, n_components=2, n_epochs=200,min_dist=0.1)
outputY2=outUmap$layout
cat("Both done","\n")
}
print("Analysis done!")
Ready for Run
We are running t-SNE
dim of inDataTsne is: 439 119
Warning: Error in .check_tsne_params(nrow(X), dims = dims, perplexity = perplexity, :
dims should be either 1, 2 or 3
{"Sample Code", "Type", "PCB-1_imputed", "PCB-2_imputed", "PCB-3_imputed", "PCB-4_10_imputed", "PCB-5_8_imputed", "PCB-6_imputed", "PCB-7_9_imputed", "PCB-11_imputed"}
"Iteration number: "
241
"Create new columns for t-SNE."
Error in eval(expr, p) : object 'outputY' not found
Cannot get R output.
Hi @MikeDereviankin, it seems like you used 4 as the Output Dim for t-SNE, which is not allowed for this version of tSNE, please use 2 or 3 instead. Alternatively, you can specify a >3 output dimension for UMAP if you think is necessary.
- Mark as Read
- Mark as New
- Bookmark
- Get Direct Link
- Report Inappropriate Content
@MJ makes a ton of sense now in hindsight based how the Rtsne algorithm works. Thanks!
- Mark as Read
- Mark as New
- Bookmark
- Get Direct Link
- Report Inappropriate Content
@MJ
Under what circumstance does the UMAP delete the rows. I understand if samples have duplicate values they will be deleted, but an UMAP identify samples with duplicate values, but different names?
@MikeDereviankin It will remove duplicate rows solely based on the predictors, even though those rows may have different names or labels. Because only predictors are used in UMAP/tSNE calculation.
Recommended Articles
- © 2026 JMP Statistical Discovery LLC. All Rights Reserved.
- Terms of Use
- Privacy Statement
- Contact Us