- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Discussions
Solve problems, and share tips and tricks with other JMP users.- JMP User Community
- :
- Discussions
- :
- Re: how to compare rows with 0 and 1 to find out the prevalence and clustering o...
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
how to compare rows with 0 and 1 to find out the prevalence and clustering of two medical conditions?
Hello, I am a new and unexperienced user of JMP working on my PhD thesis. I simply cannot find out how to compare a part of my data and I need assistance. My data consists of 146 columns with either 1 or 0 (yes/no) for 146 medical conditions respectively. I have about 14000 rows (patients) to compare this data from. I need to find out the order for the most common combinations for two medical conditions in the same person. How can I do that?
very grateful for help,
Emma
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: how to compare rows with 0 and 1 to find out the prevalence and clustering of two medical conditions?
Hi @EmmaLT ,
I think I have an idea of how to do what you're asking about, but it would help if you could share some of your data -- you can anonymize it if need be. It doesn't need to be the whole data set, just a subset so that I can better understand how the data is structured as that might impact how to do the analysis.
DS
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: how to compare rows with 0 and 1 to find out the prevalence and clustering of two medical conditions?
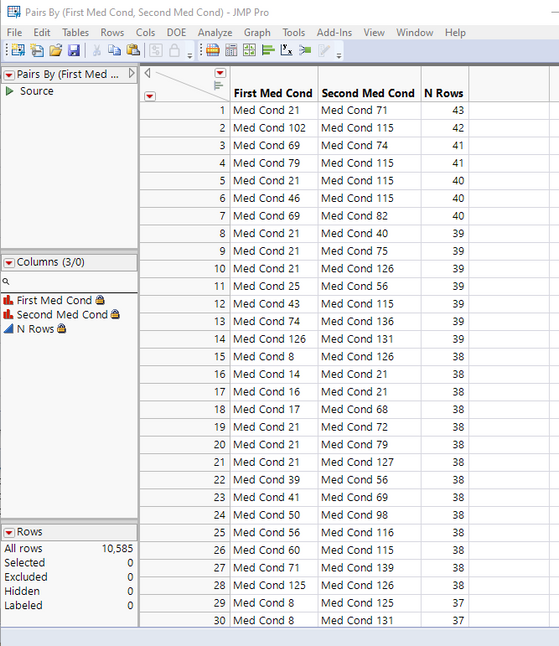
I have created a sample script, that generates an example data table to run the analysis on. I only am creating 100 rows of data because it takes a while to create the pairs of data. The resulting table provides the count of the number of patients each combination of medical conditions were found in.
The script will take a good bit of time if you run it on all 14000 patients at once. You can easily run it on subsets of the data, and then combine the resulting tables.
Names Default To Here( 1 );
dt = New Table( "raw Data" );
dt << add rows( 100 );
dt << New Column( "Patient", set each value( Row() ) );
Try( dt << delete columns( "Column 1" ) );
For( i = 1, i <= 146, i++,
dt << New Column( "Med Cond " || Char( i ),
set each value( Round( Random Uniform( 0, 1 ), 0 ) )
)
);
// Create the paired data table
dtPair = New Table( "Pairs",
New Column( "Patient" ),
New Column( "First Med Cond", Character ),
New Column( "Second Med Cond", Character )
);
//Populate the Pairs Table
For( theRow = 1, theRow <= N Rows( dt ), theRow++,
For( theFirstCond = 2, theFirstCond <= N Cols( dt ), theFirstCond++,
If( Column( dt, theFirstCond )[theRow] == 1,
thePatient = dt:Patient[theRow];
firstCond = (Column( dt, theFirstCond )) << get name;
For( theSecondCond = theFirstCond + 1, theSecondCond <= N Cols( dt ),
theSecondCond++,
If( Column( dt, theSecondCond )[theRow] == 1,
dtPair << add row( 1 );
pairRow = N Rows( dtPair );
dtPair:Patient[pairRow] = thePatient;
dtPair:First Med Cond[pairRow] = firstCond;
dtPair:Second Med Cond[pairRow] = (Column( dt, theSecondCond )) << get name;
)
);
)
)
);
dtSummary = dtPair << Summary(
Group( :First Med Cond, :Second Med Cond ),
Freq( "None" ),
Weight( "None" )
);
dtSummary << sort( by(N Rows), order(descending), replace table(11));- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: how to compare rows with 0 and 1 to find out the prevalence and clustering of two medical conditions?
Thank you txnelson and all others for your advice and suggestions! I came to a halt within the analysis and had to start over with the extraction of the original data, so i haven't been able to try out the solutions yet. I will be back when it's time. Thank you!
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: how to compare rows with 0 and 1 to find out the prevalence and clustering of two medical conditions?
It occurs that "the order for the most common combinations for two medical conditions in the same person" could imply a few different analysis objectives. But, whatever analysis is implied, I would strongly advocate taking a look at what's 'in' your data first.
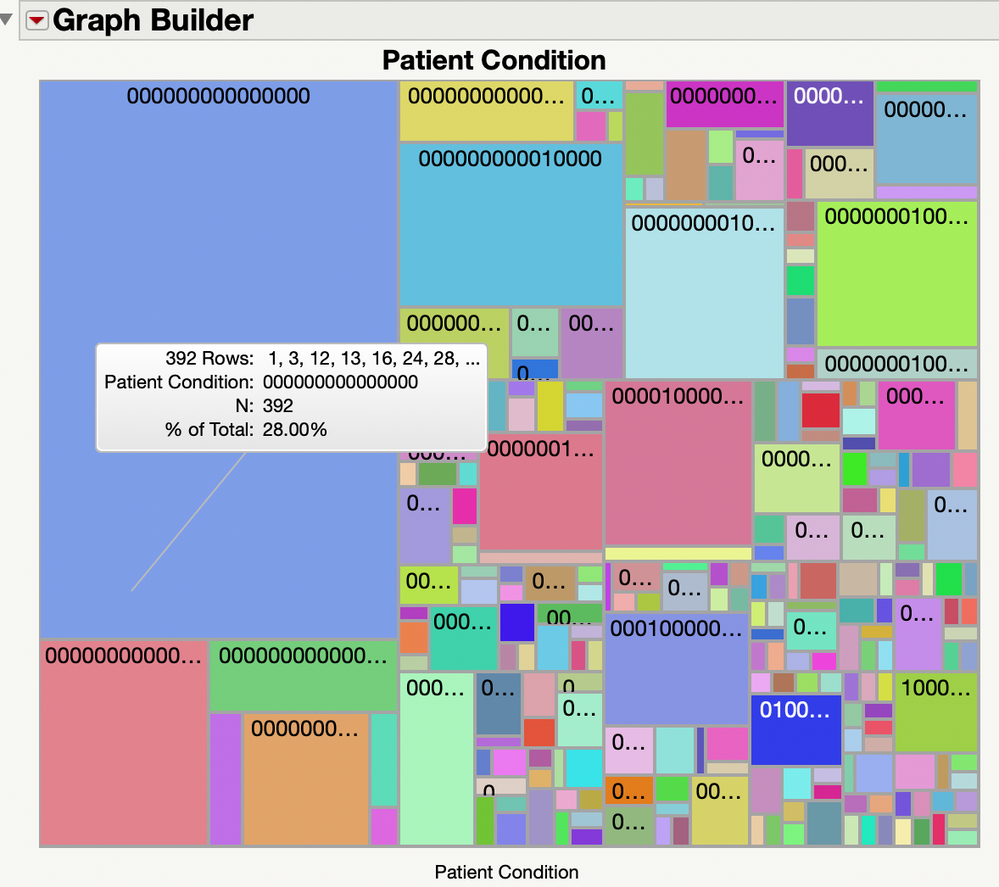
One approach would be to use "Cols > Utilities > Combine Columns...' to make a new column containing the status of each patient for every condition. Then you can use 'Graph > Graph Builder' to make a Treemap of this new column, and inspect the results via the tooltip. If you are not sure of the details of how to do this, try the online documentation. Be aware that, with so many conditions, this may just be a mess.
As a new user, you probably are not interested in scripting. But the script below simulates some data, and makes the Treemap. Because of the randomness, what you see may be different each time. But running this code should produce a graph like:
To run the script do 'File > New >New Script', paste the code into the window that appears, then do 'Edit > Run Script'.
NamesDefaultToHere(1);
// Generate some fake binary data for each condition (maximum possible incidence for one condition is 20% of the patients):
// This is unrepresentative since the incidences of the conditions are treated as independent, but the in practice this may not
// be the case
nr = 1400; // Number of patients
nc = 15; // Number of conditions
m = J(nr, nc, 0);
for(c=1, c<=nc, c++,
incidence = RandomInteger(0, Round(0.2 * nr));
m[RandomIndex(nr, incidence), c] = 1;
);
dt = asTable(m);
dt << setName("Fake Incidence Data");
// Use 'Combine Columns' to make a binary string for each patient indicating what conditions they have
cols = dt << getColumnNames;
dt << combineColumns(columns(Eval(cols)), ColumnName("Patient Condition"), Delimiter( "" ), MultipleResponse(0));
// Make a Treemap of the patient conditions
dt << Graph Builder(
Show Control Panel( 0 ),
Show Legend( 0 ),
Variables( X( :Patient Condition ) ),
Elements( Treemap( X, Legend( 1 ) ) )
);
// Aggregate the patient comnditions to a new table and sort it (descending)
//dt2 = dt << Summary(Group(:Patient Condition));
//dt2 << Sort(By(:N Rows), ReplaceTable, Order(Descending));
Recommended Articles
- © 2026 JMP Statistical Discovery LLC. All Rights Reserved.
- Terms of Use
- Privacy Statement
- Contact Us