- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Discussions
Solve problems, and share tips and tricks with other JMP users.- JMP User Community
- :
- Discussions
- :
- Re: R interface in JMP
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
R interface in JMP
Hello,
I wrote a code in R that I want to run into JMP. Here is the code :
R init();
//send data to R
R Send(dt);
R submit("
library(spatstat)
#creation fenetre
fenn<-owin(xrange=c(40,760), yrange=c(40,160), poly=NULL, mask=NULL, units=NULL)
ppp<-ppp(x=dt$XM,y=dt$YM,window=fenn)
print(quadratcount(ppp,nx=6,ny=3)
Q<-quadrat.test.ppp(ppp,nx=6,ny=3)
a<-cdf.test(ppp,'x')
D=0
if(a[[2]] & Q[[3]] < 0.1 ){
D=0}else {D=1}
Distribution=as.data.frame(rep(c(D),nrow(dt)))
class(Distribution)
print(a)
");
R term();
Then when I run it I got two issues:
Firstly I have an error from these lines :
fenn<-owin(xrange=c(40,760), yrange=c(40,160), poly=NULL, mask=NULL, units=NULL)
ppp<-ppp(x=dt$XM,y=dt$YM,window=fenn)which says :
Error: Function boundingbox called with 2 unrecognised arguments
In addition: Warning message:
2 points were rejected as lying outside the specified window
In fact when I change 40 into 0 it works because all my points are in my window, but when I run the code on R the fact that all my points are not in the window is not an issue. And if I run the previous code in R it works. So I don't understand why I got a warning message in JMP but not in R.
The second issue is when I want to export my data, JMP don't want to extract my data frame and I don't understand why. I use this code :
ResidJMP = R Get(Distribution);
ResidJMP << New Data View;
R term();In fact I want to return a data table in JMP and after I want to use it as the main data table.
- Tags:
- windows
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: R interface in JMP
Hello,
For a basic example,
R Init(); // Initiate R connection
dt = Open(“$SAMPLE_DATA/Big Class.jmp”,invisible);
R Send(dt);the call of column should be done on the following way:
R Submit("
print(dt['weight'])
//b<-dt['weight']/dt['height']
");
instead of
R Submit("
print(dt$weight)
//b<-dt['weight']/dt['height']
");I think it should change the output of your first problem.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: R interface in JMP
When I apply this to my data it says that the columns I used aren't numeric, then when I use as.numeric() function it says : 'list' object cannot be coerced to type 'double'. This problem doesn't appeared with the $ call. There are so many issues here unfortunately
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: R interface in JMP
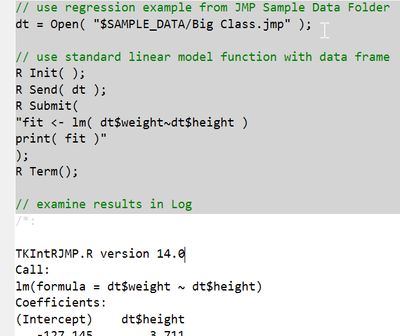
You can use a JMP data table as a data frame in R. The R Send() function automatically converts one data structure into the other. Here is a simple example:
Names Default to Here( 1 );
// make sure JMP can find the desired installed version of R
Set Environment Variable( "R_HOME", "C:\Program Files\R\R-4.1.3" );
// use regression example from JMP Sample Data Folder
dt = Open( "$SAMPLE_DATA/Big Class.jmp" );
// use standard linear model function with data frame
R Init( );
R Send( dt );
R Submit(
"fit <- lm( dt$weight~dt$height )
print( fit )"
);
R Term();
// examine results in Log
So I suspect that there is a problem with the R code (function arguments), or the nature of the variables in the data frame. After you make sure that your function call is correct, try printing the data table from R to see if everything transferred as expected.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: R interface in JMP
When I use this code on R
library(spatstat)
data<-read.("Results.csv")
#creation fenetre
fenn<-owin(xrange=c(40,760), yrange=c(40,160), poly=NULL, mask=NULL, units=NULL)
#creation point pattern
ppp<-ppp(x=data$XM,y=data$YM,window=fenn)
plot(ppp)
summary(ppp)
#test quadrat
quadratcount(ppp,nx=6,ny=3)
Q<-quadrat.test.ppp(ppp,nx=6,ny=3)
paste("Ma pvalue du quadrat test ", format(Q[[3]], scientific=TRUE, digits=3))
#test ks
a<-kstest(ppp,'x')
paste("Ma pvalue du KS test ", format(a[[2]], scientific=TRUE, digits=3))
D=character()
if(a[[2]] & Q[[3]] < 0.1 ){
D="Distribution Non-Uniforme"}else {D="Distribution Uniforme"}
Distribution<-rep(c(D),length(XM))
dt<-cbind(dt,Distribution)
It works good. In fact it works good in JMP if I use this window :
fenn<-owin(xrange=c(0,760), yrange=c(0,160), poly=NULL, mask=NULL, units=NULL)
Because all my (x,y) points are in the window, the problem comes out when I put the window I want. But in R there is no problem of window.
But if there is a problem with the variables it shouldn't work with an other window and this is what I don't understand.
Maybe there is something I don't understand in your comment but I don't find what could be the error. When I print it on JMP from R I have the good data frame. I'm lost for sure
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: R interface in JMP
Did you try my simple example? Does it work? What do you see in the JMP Log?
Did you try sending a JMP data table to R and printing it in R? Does that data frame match the data table?
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: R interface in JMP
Your example works I had this in the log :
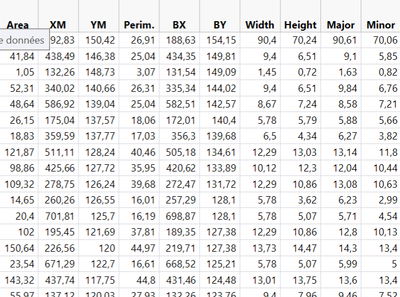
For your second question I'm not sure I'm understanding it well. But I try to send my JMP data table to R with send() command and then print it in the log. For your example it looks like this :
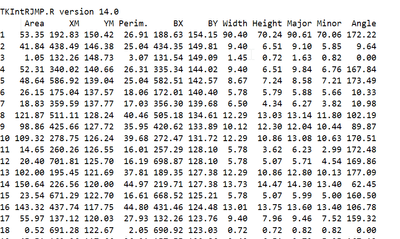
In my example i got this in the log :
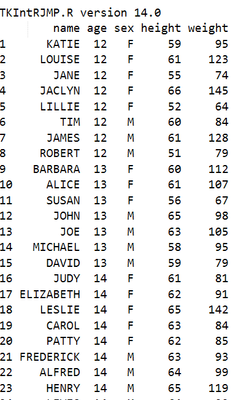
and my table look like this :
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: R interface in JMP
Now my main problem is to export my data frame from R. I have the same error with your example than with my data.
Erreur : Error in if (is.na(datam)) { : the condition has length > 1
Unexpected errors occurred while attempting to transfer the data.I check that my R get() is on a data.frame. Here is my code :
Treat=Open( "C:\Users\GROUSD01\Desktop\CréationcodeMPPT\Results 4.jmp" );
R init();
//send data to R
R Send(Treat);
R submit("library(spatstat)
round(Treat$XM)
round(Treat$YM)
XM<-Treat$XM
YM<-Treat$YM
test<-as.data.frame(cbind(XM,YM))
test2<-data.frame()
for (i in 1:nrow(test)){
if(test[i,2]>39){
test2<-rbind(test2,test[i,1:2])}}
fenn<-owin(xrange=c(40,760), yrange=c(40,160), poly=NULL, mask=NULL, units=NULL)
#creation point pattern
ppp<-ppp(x=test2$XM,y=test2$YM,window=fenn)
print(ppp)
plot(quadratcount(ppp,nx=6,ny=3))
Q<-quadrat.test.ppp(ppp,nx=6,ny=3)
print(Q)
a<-cdf.test(ppp,'x')
paste('Ma pvalue du KS test ', format(a[[2]], digits=3))
D=character()
if(a[[2]] & Q[[3]] < 0.1 ){
D='Distribution Non-Uniforme'}else {D='Distribution Uniforme'}
Distribution<-(rep(c(D),length(Treat$XM)))
Treat<-cbind(Treat,Distribution)
");
R Get(Treat)<<New Data View
R term();But I tried a lot of different things for the same results
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: R interface in JMP
I suggest that you contact JMP Technical Support (support@jmp.com) for help with this problem.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: R interface in JMP
Was there a solution to the error?
Error in if (is.na(datam)) { : the condition has length > 1I have the same issue.
Recommended Articles
- © 2026 JMP Statistical Discovery LLC. All Rights Reserved.
- Terms of Use
- Privacy Statement
- Contact Us