- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Discussions
Solve problems, and share tips and tricks with other JMP users.- JMP User Community
- :
- Discussions
- :
- Re: Keyword cooccurence networks in JMP?
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Keyword cooccurence networks in JMP?
Accepted Solutions
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Keyword cooccurence networks in JMP?
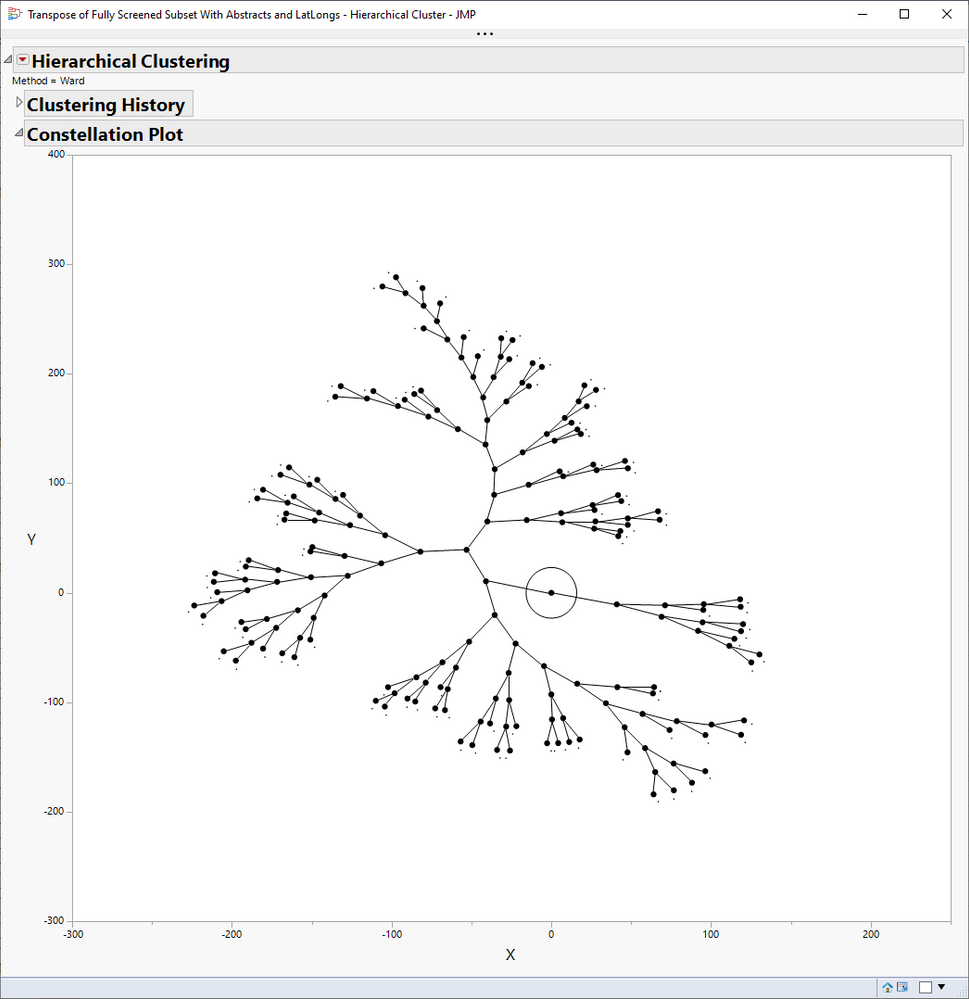
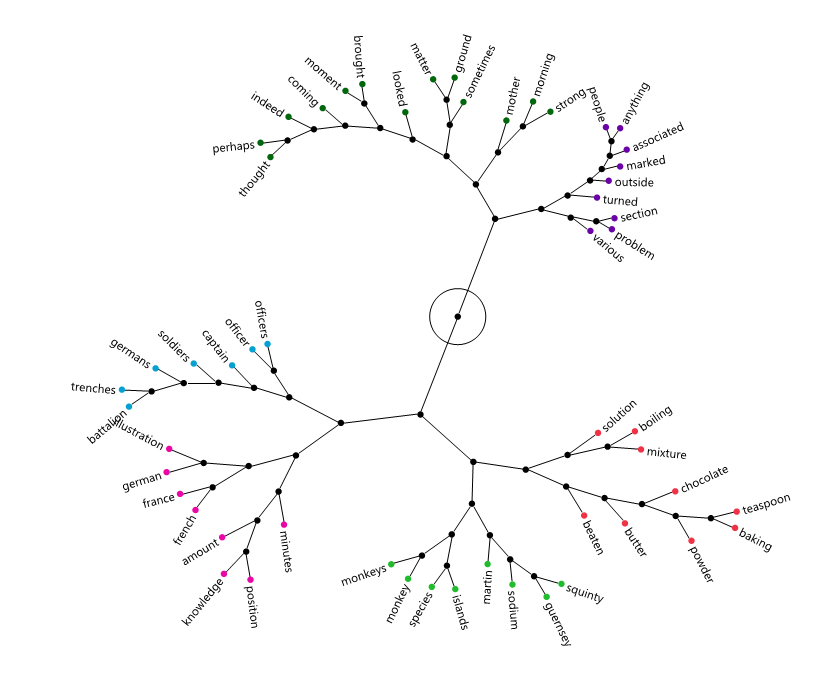
This isn't really it either, but maybe closer. I colored these by selecting a root node in the branch and then coloring the selected rows in the data table. It took a bit of hand-curating the stop words to make a pretty picture.
// load some documents that might separate into some categories
dt1 = Open( "f:/gutenberg/books5000.jmp" ); // 5000 is too many, subset 36...
dt1 << selectwhere(
Starts With( Loc Class, "D501" ) | Starts With( Loc Class, "TX:" ) | Starts With( Loc Class, "Q" ) | Starts With( Loc Class, "P" )
);
dt2 = dt1 << subset( selected rows( 1 ) );
Close( dt1, nosave );
originalnames = dt2 << getColumnNames();
te = dt2 << Text Explorer(
Text Columns( :text ), // the entire document is in one cell of the row
Add Stop Words(
{"agreement", "almost", "another", "company", "copyright holder", "electronic", "foundation", "gutenberg", "literary archive", "little",
"person or entity", "project", "public domain", "research", "without", "ebooks", "enough", "though", "rather", "better", "common", "possible",
"weight", "present", "series", "necessary", "placed", "therefore", "towards", "footnote", "something","slowly","around","behind","looking","seemed","nothing",
"probably","called","easily","distributing","paragraph"
}
),
Minimum Characters per Word( 6 ),
Stemming( "no stemming" ), //"Stem for Combining"
Language( "English" )
);
//te << savedocumenttermmatrix( Maximum Number of Terms( 300 ), Minimum Term Frequency( 25 ), Weighting( "TF IDF" ) );//BINARY would use ALL rather than !any, below
te << savedocumenttermmatrix( Maximum Number of Terms( 100 ), Minimum Term Frequency( 25 ), Weighting( "BINARY" ) ); // 0 or 1 if it occurs
te << closewindow;
// remove all-connected columns
allnames = dt2 << getColumnNames();
For( iname = N Items( allnames ), iname > N Items( originalnames ), iname -= 1,
// If( !Any( dt2[0, iname] ),
If( All( dt2[0, iname] ),
dt2 << deletecolumns( iname )
)
);
allnames = dt2 << getColumnNames();
cols = (N Items( originalnames ) + 1) :: N Items( allnames );
//
//dt2 << Hierarchical Cluster( Y( allnames[cols] ),
// Label( Transform Column( "Label", Character, Formula( Left( left(LoC Class,4)||:Subject||:Title, 20 ) ) ) ), // build your own identifier here
// Method( "Ward" ), Standardize Data( 1 ), Dendrogram Scale( "Distance Scale" ),
// Number of Clusters( 4 ), Constellation Plot( 1 ), Show Dendrogram( 0 ),
// SendToReport(Dispatch({"Constellation Plot"},"Clust Hier",FrameBox,{Frame Size( 1056, 716 )}))
//);
dt3 = dt2 << Data Table( "Subset of books5000" ) << Transpose(
columns( allnames[cols] ),
Label( :Title ),
Label column name( "Title" ),
Output Table( "Transpose of Subset of books5000" )
);
dt3 << Hierarchical Cluster(
Y( (dt3 << getColumnNames)[2 :: N Cols( dt3 )] ),
Label( Transform Column( "Transform[Title]", Nominal, Formula( Left( :Title, Length( :Title ) - 7 ) ) ) ),
Method( "Ward" ),
Standardize Data( 1 ),
Show Dendrogram( 0 ),
Dendrogram Scale( "Distance Scale" ),
Number of Clusters( 13 ),
Constellation Plot( 1 ),
SendToReport( Dispatch( {"Constellation Plot"}, "Clust Hier", FrameBox, {Frame Size( 948, 827 )} ) )
);- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
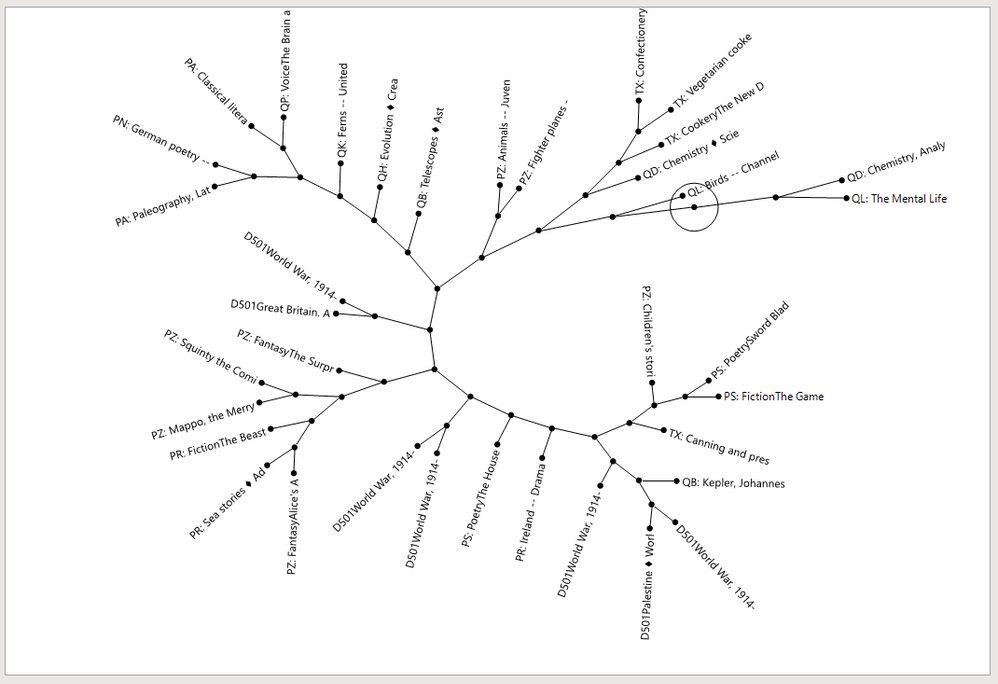
Re: Keyword cooccurence networks in JMP?
You can get something related, maybe, like this. And it is connected to the table. And not Pro.
// load some documents that might separate into some categories
dt1 = Open( "f:/gutenberg/books5000.jmp" ); // 5000 is too many, subset 36...
dt1 << selectwhere( Starts With( Loc Class, "D501" ) | Starts With( Loc Class, "TX:" ) | Starts With( Loc Class, "Q" ) | Starts With( Loc Class, "P" ) );
dt2 = dt1 << subset( selected rows( 1 ) );
Close( dt1, nosave );
originalnames = dt2 << getColumnNames();
te = dt2 << Text Explorer(
Text Columns( :text ), // the entire document is in one cell of the row
Add Stop Words(
{"agreement", "almost", "another", "company", "copyright holder", "electronic", "foundation", "gutenberg", "literary archive", "little",
"person or entity", "project", "public domain", "research", "without"}
),
Minimum Characters per Word( 6 ),
Stemming( "Stem for Combining" ),
Language( "English" )
);
te << savedocumenttermmatrix( Maximum Number of Terms( 300 ), Minimum Term Frequency( 25 ), Weighting( "TF IDF" ) );//BINARY would use ALL rather than !any, below
te << closewindow;
// remove all-connected columns
allnames = dt2 << getColumnNames();
For( iname = N Items( allnames ), iname > N Items( originalnames ), iname -= 1,
If( !Any( dt2[0, iname] ),
dt2 << deletecolumns( iname )
)
);
allnames = dt2 << getColumnNames();
cols = (N Items( originalnames ) + 1) :: N Items( allnames );
dt2 << Hierarchical Cluster( Y( allnames[cols] ),
Label( Transform Column( "Label", Character, Formula( Left( left(LoC Class,4)||:Subject||:Title, 20 ) ) ) ), // build your own identifier here
Method( "Ward" ), Standardize Data( 1 ), Dendrogram Scale( "Distance Scale" ),
Number of Clusters( 4 ), Constellation Plot( 1 ), Show Dendrogram( 0 ),
SendToReport(Dispatch({"Constellation Plot"},"Clust Hier",FrameBox,{Frame Size( 1056, 716 )}))
);- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Keyword cooccurence networks in JMP?
Hi Craige,
This is brilliant! I'm not a JSL whiz, but I got it working with my data. The only thing I can't figure out how to do is to create a label identifier that uses the column names for terms in the document term matrix. My document term matrix has 252 columns. Each column name is autoformatted with the terms stemmed value and the suffix TF IDF2. For example, one column is named "distribut· TF IDF 2" another is named "mountain· TF IDF 2". I'd like to have the constellation point labels be the stemmed term, without the suffix (e.g., distribut· or mountain·). Is there an easy way to use the values from the column names of the document term matrix as point labels?
Thank you so much for providing this script.
Casey
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Keyword cooccurence networks in JMP?
I'm not sure; I think for each row you would want the 3 or 4 most important words (column names) for that row, and they might be the ones with the biggest value. Does that look right when you look at some rows? If so, it would not be much of a preprocessing step to create that label column.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Keyword cooccurence networks in JMP?
Using one row at a time may not be the right answer; that won't have to be a common word(s) with neighbors in the diagram. Unless two spatial neighbors share some words.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Keyword cooccurence networks in JMP?
Hi Craige,
I just realized that the script you sent uses values in a DTM to create clusters of documents based on similar uses of terms among documents. In this case, each node represents a document and each link is based on a distance value from cluster analysis. I'm trying to do something slightly different.
I'd like to create a constellation graph like this where each node represents a TERM in the DTM and each link is based on some sort of statistic that describes keyword co-occurrence. This is way outside of my expertise. I love the idea of such a plot, but I'm not sure if it would be appropriate to reformat the DTM somehow so that a cluster analysis could be done where terms end up as nodes (or how I would do this). This would accomplish my goal of having a graphic in JMP based on term relatedness that I can use as a global filter in an interactive data visualization. However, would this sort of end-around create a graphic that is not based on best practices of text analysis? For those of you that understand this field, would this use of cluster analysis be inappropriate to illustrate keyword co-occurrence? If so, has anyone built anything that can do keyword cooccurrence analyses (using proper statistical methods for this type of analysis) in JMP? I'm interested in hearing feedback from the community on this. I apologize that my original post wasn't clearer. Craige, I will definitely use the script that you sent in your first response for when I need to cluster documents! Thank you.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Keyword cooccurence networks in JMP?
Just adding to @Craige_Hales' help that you can use Latent Class Analysis to group rows (documents) and Latent Semantic Analysis to group columns (terms). These platforms are available under Analyze > Multivariate Methods. (They are also built in and specialized for Text Explorer in JMP Pro.) See JMP documentation...
The idea is that JMP has a lot of powerful platforms that work together. My analogy is a graphing calculator. There are a lot of buttons and it often takes more than one button to get the job done. That is to say that there is no one button that 'does it all,' but you can get there. You need to learn JMP as the first approach instead of trying to re-create a custom application. Why re-invent the wheel?
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Keyword cooccurence networks in JMP?
Hi @Mark_Bailey,
I've been a JMP loyalist since 1994, so I'm aware of it's many strengths and how different problems can be solved via different pathways (that's just one of things I love about it). I always start with JMP. My professional community is dominated by R users. They can do all kinds of analyses, but R's ability to create interactive data visualizations (my thing) are not even in the same league as JMP. So, I stick with JMP. However, as statistical methods evolve, a greater fraction of newer analyses are possible only through R, or through JMP Pro, but not using a standard JMP license (e.g., various text analysis platforms, basic model selection procedures that are standard practice in my field). This makes it harder for me to do holistic, start to finish analyses in JMP. As JMP Pro and standard JMP drift farther apart, I find myself consulting the community for possible solutions to my problems that I can pull of with a standard JMP license. Craige's post was a great example of the community helping me find a way to group documents using a DTM in standard JMP. My most recent post asked if there is a similar solution that may be possible to group terms using a DTM in standard JMP.
In this specific case, I'm trying to incorporate a keyword co-occurrence network graphic into an existing interactive visualization that I have created in standard JMP. I don't think I'm trying to re-invent the wheel. I think I'm either running into a wall of functionality in the standard JMP license (maybe) or a lack of knowledge/ creativity on my part on how to pull this off (more likely). I can't afford JMP Pro. This is my reality and I don't see it changing any time soon. Both the Latent Class Analysis and Latent Semantic analysis platforms you mentioned are only available in JMP Pro. When I ask questions about capabilities of the standard JMP license and get responses telling me that I can only do something in JMP Pro, it doesn't really help me. I'm happy if my questions help solve problems for JMP Pro users, but I'm still left with my original conundrum of trying to do something in standard JMP that requires more creativity and knowledge (two amazing qualities of the JMP user community) than I possess on my own.
I'm grateful for JMP, for JMP's technical support staff (who are amazing), and for this user community. Until my budget becomes way bigger than it's ever been, I'll probably keep asking questions here about creative ways to get things done using a standard JMP license (that may be pushing the envelope at times). I agree with Mark that one needs to learn JMP first (which I have enjoyed doing for over 25 years). Still, I struggle at times to understand how to meet my expanding analysis and visualization needs with a standard JMP license. I'm still hoping there is a legitimate way to use a standard JMP to create a keyword co-occurrence network diagram to group terms that has full interactivity and can be saved as a JMP table script. I don't know how to do this. Given my normal process of reading JMP documentation, searching JMP community posts, and just experimenting on my own, I haven't been able to figure out how to do this. If it just can't be done without a JMP Pro license, I'll let it drift and leave this graph type out of my visualization.
Sorry for the long response. If anyone from JMP wants to weigh in with some strategies on how to get by with standard JMP when you can't afford JMP Pro, I'd love some advice (seems like a good blog topic). Perhaps it's not a topic for this discussion board. I apologize if it isn't. Feel free to contact me offline at caseylott@ymail.com if anyone has suggestions for this.
Casey
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Keyword cooccurence networks in JMP?
This isn't really it either, but maybe closer. I colored these by selecting a root node in the branch and then coloring the selected rows in the data table. It took a bit of hand-curating the stop words to make a pretty picture.

// load some documents that might separate into some categories
dt1 = Open( "f:/gutenberg/books5000.jmp" ); // 5000 is too many, subset 36...
dt1 << selectwhere(
Starts With( Loc Class, "D501" ) | Starts With( Loc Class, "TX:" ) | Starts With( Loc Class, "Q" ) | Starts With( Loc Class, "P" )
);
dt2 = dt1 << subset( selected rows( 1 ) );
Close( dt1, nosave );
originalnames = dt2 << getColumnNames();
te = dt2 << Text Explorer(
Text Columns( :text ), // the entire document is in one cell of the row
Add Stop Words(
{"agreement", "almost", "another", "company", "copyright holder", "electronic", "foundation", "gutenberg", "literary archive", "little",
"person or entity", "project", "public domain", "research", "without", "ebooks", "enough", "though", "rather", "better", "common", "possible",
"weight", "present", "series", "necessary", "placed", "therefore", "towards", "footnote", "something","slowly","around","behind","looking","seemed","nothing",
"probably","called","easily","distributing","paragraph"
}
),
Minimum Characters per Word( 6 ),
Stemming( "no stemming" ), //"Stem for Combining"
Language( "English" )
);
//te << savedocumenttermmatrix( Maximum Number of Terms( 300 ), Minimum Term Frequency( 25 ), Weighting( "TF IDF" ) );//BINARY would use ALL rather than !any, below
te << savedocumenttermmatrix( Maximum Number of Terms( 100 ), Minimum Term Frequency( 25 ), Weighting( "BINARY" ) ); // 0 or 1 if it occurs
te << closewindow;
// remove all-connected columns
allnames = dt2 << getColumnNames();
For( iname = N Items( allnames ), iname > N Items( originalnames ), iname -= 1,
// If( !Any( dt2[0, iname] ),
If( All( dt2[0, iname] ),
dt2 << deletecolumns( iname )
)
);
allnames = dt2 << getColumnNames();
cols = (N Items( originalnames ) + 1) :: N Items( allnames );
//
//dt2 << Hierarchical Cluster( Y( allnames[cols] ),
// Label( Transform Column( "Label", Character, Formula( Left( left(LoC Class,4)||:Subject||:Title, 20 ) ) ) ), // build your own identifier here
// Method( "Ward" ), Standardize Data( 1 ), Dendrogram Scale( "Distance Scale" ),
// Number of Clusters( 4 ), Constellation Plot( 1 ), Show Dendrogram( 0 ),
// SendToReport(Dispatch({"Constellation Plot"},"Clust Hier",FrameBox,{Frame Size( 1056, 716 )}))
//);
dt3 = dt2 << Data Table( "Subset of books5000" ) << Transpose(
columns( allnames[cols] ),
Label( :Title ),
Label column name( "Title" ),
Output Table( "Transpose of Subset of books5000" )
);
dt3 << Hierarchical Cluster(
Y( (dt3 << getColumnNames)[2 :: N Cols( dt3 )] ),
Label( Transform Column( "Transform[Title]", Nominal, Formula( Left( :Title, Length( :Title ) - 7 ) ) ) ),
Method( "Ward" ),
Standardize Data( 1 ),
Show Dendrogram( 0 ),
Dendrogram Scale( "Distance Scale" ),
Number of Clusters( 13 ),
Constellation Plot( 1 ),
SendToReport( Dispatch( {"Constellation Plot"}, "Clust Hier", FrameBox, {Frame Size( 948, 827 )} ) )
);- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Keyword cooccurence networks in JMP?
Hi Craige,
That's it! I modified the script you sent for my data table (see attached script and table file). Everything worked great, but I couldn't get the label function to work. I looked at it 3 times and I swear I'm copying your code exactly. Do you see anything that could be keeping it from working? The plot image that I get is below. As you can see, the term labels are shown only as dots. Thank you again. I really appreciate it!
Recommended Articles
- © 2026 JMP Statistical Discovery LLC. All Rights Reserved.
- Terms of Use
- Privacy Statement
- Contact Us