- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Discussions
Solve problems, and share tips and tricks with other JMP users.- JMP User Community
- :
- Discussions
- :
- Import x and y values from the text file to jmp table
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Import x and y values from the text file to jmp table
Hi,
Reading each line in text file and import each string as a separate row in the jmp data table
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Import x and y values from the text file to jmp table and create wafer map
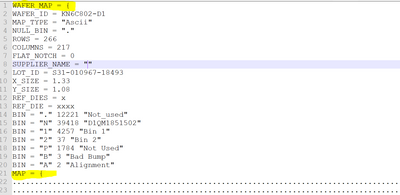
There are two strings containing "MAP = {" highlighted below:
I tried using & statement to not select first one but it doesn't work.
demo_str = Load text file("New wafer map.txt");
a = Contains(demo_str,"MAP = {") & Contains(demo_str,"WAFER_MAP = {") ==0; // get the location of the string "MAP ={"
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Import x and y values from the text file to jmp table and create wafer map
You can use Contains to skip first "MAP = {" or search from end (read scripting index...)
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Import x and y values from the text file to jmp table and create wafer map
Hope you are doing well @jthi,!
Real quick question on this discussion-
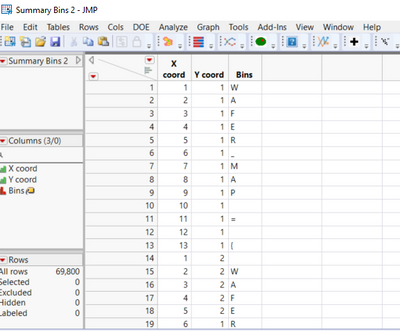
I would also like to import all the strings from the txt file in the jmp data table including the header information. The final data table should contain all the strings and each character should be treated as a separate word. It should look like this.
The logic that you suggested doesn't work after row 13. Any help would be much appreciated
map_str =
"\[
WAFER_MAP = {
WAFER_ID = ""
BIN G = "Passing"
MAP = {
.....GGG.....
....BGGGG....
...BGGGGGG...
..GGGGGGGGG..
.GGGGGGBBGGG.
GGGGGGGBBGGGB
GGGGGGGGGGGGB
GGGGGGGGGGGGB
.GGGGGGGGGGG.
..GGGGGGGGG..
...GGGGGGG...
....GGGGG....
.....GGG.....
}
}
]\";
l = Words( map_str, "\!N" );
rowc = N Items( l );
colc = N Items( Words( l[1], "" ) );
dt = New Table( "Summary Bins",
Add Rows( rowc * colc ),
New Column( "X coord", Numeric, Ordinal ),
New Column( "Y coord", Numeric, Ordinal ),
New Column( "Bins", Character, Nominal, Label( 1 ) )
);
Try(
For Each( {line, idx}, l,
cur_rows = ((idx - 1) * colc :: (idx * colc - 1)) + 1;
dt[cur_rows, "X coord"] = ((1 :: colc))`;
;
dt[cur_rows, "Y coord"] = (J( 1, colc ) * (idx))`;
dt[cur_rows, "Bins"] = Words( line, "" );
)
);Thanks for your time!
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Import x and y values from the text file to jmp table and create wafer map
I would change method if you want the other information too. As the data in the file has new data on each line (except for wafermap) I would use Words() and split on row change (you can still look for when wafermap starts by using Contains from the list Words() produces, but it is index in a list, not in string). Then I would add the information to associative array to make accessing it easier (this will require some looping and more Words() usage))
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Import x and y values from the text file to jmp table and create wafer map
Thank Jamo @jthi for letting me know what I was missing. I knew I was close. Can you share a example code just so I can refer? I am still learning how to play with strings
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Import x and y values from the text file to jmp table and create wafer map
Again, reminder to use scripting index and jmp help. They are usually best resources to learn this type of JMP Scripting
Names Default To Here(1);
example_str = "\[
WAFER_MAP = {
WAFER_ID = "ABC"
A B C = 123
]\";
example_list = Words(example_str, "\!N");
Show(example_list);
Remove From(example_list, 1); // drop first index. Why it is fine in this case?
// loop over lines until "MAP = {" I'll just use hardcoded index for example
// I'll also save values to variables
// but associative array would be better in this case
wafer_id_line = example_list[1];
Show(wafer_id_line);
wafer_id = Word(2, wafer_id_line, "="); // you might have to remove extra quotes
show(wafer_id);
example_aa = Associative Array();
//adding values to associative array could be done with something like
example_aa["WAFER_ID"] = Word(2, wafer_id_line, "="); // you might have to remove extra quotes
show(example_aa);
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Import x and y values from the text file to jmp table and create wafer map
Here is complete example for this which uses associative array to store information about the wafer and builds jmp table from the text file.
Names Default To Here(1);
// jthi 2022-08-02
// https://community.jmp.com/t5/Discussions/Import-x-and-y-values-from-the-text-file-to-jmp-table-and-create/m-p/518658
// Load text file to variable
txt = Load Text File("$DOWNLOADS/wafer map 1.txt");
// convert to list of lines
lines = Words(txt, "\!N");
// drop first and last two lines
Remove From(lines, 1);
Remove From(lines, N Items(lines));
Remove From(lines, N Items(lines));
// separate MAP to different variable
map_start_idx = Contains(lines, "MAP = {");
map_lines = Remove From(lines, map_start_idx, N Items(lines) - map_start_idx + 1);
Remove From(map_lines, 1);
// associative array to collect wafer info
aa_info = Associative Array();
aa_info["BINS"] = Associative Array("");
bin_rgx_pattern = "\[^"(.+)" (\d+) "(.+)"$]\";
For Each({line}, lines,
key = Trim Whitespace(Word(1, line, "="));
value = Trim Whitespace(Word(2, line, "="));
If(key == "BIN",
new_bin = Associative Array({"count", "description"}, "");
bin_list = Regex Match(value, bin_rgx_pattern);
new_bin["count"] = bin_list[3];
new_bin["description"] = bin_list[4];
aa_info["BINS"][bin_list[2]] = new_bin;
,
value = Trim Whitespace(Word(2, line, "="));
aa_info[key] = Substitute(value, "\!"", "");
);
);
// create wafermap data table
rowc = N Items(map_lines);
colc = N Items(Words(map_lines[1], ""));
dt = New Table(aa_info["WAFER_ID"],
Add Rows(rowc * colc),
New Column("X", Numeric, Continuous),
New Column("Y", Numeric, Continuous),
New Column("bin", Character, Nominal, Label(1)),
invisible
);
For Each({line, idx}, map_lines,
cur_rows = ((idx - 1) * colc::(idx * colc - 1)) + 1;
dt[cur_rows, "X"] = ((1::colc))`;
dt[cur_rows, "Y"] = (J(1, colc) * (idx))`;
dt[cur_rows, "bin"] = Words(line, "");
);
// at this point we can use aa_info to set values different places: table variable, column properties, cells, ...
dt << New Table Variable("WAFER_INFO", char(aa_info)); // usage Parse(:WAFER_INFO)["BINS"]["."]
dt << New Column("BIN_COUNT", Numeric, Continuous, << Set Each Value(Col Sum(1, :bin)));
dt << New Column("BIN_COUNT_SANITY", Numeric, Continuous, << Set Each Value(Num(aa_info["BINS"][:bin]["count"])));
// use value labels for bins column
bin_labels = {};
For Each({{key, value}}, aa_info["BINS"],
Insert Into(bin_labels, Parse(Eval Insert("\["^key^" = "^value["description"]^"]\")));
);
// Seems like JMP won't apply value label to "." correctly
Column(dt, "bin") << Value Labels(Eval(bin_labels)) << Use Value Labels(1);
// or add description column
dt << New Column("BIN_DESC", Character, Nominal, << Set Each Value(aa_info["BINS"][:bin]["description"]));
// we can set null_bin as missing value code column proeprty
Column(dt, "bin") << Set Property("Missing Value Codes", (aa_info["NULL_BIN"]));
dt << Show Window(1);
show(aa_info);
Write();I did also notice that seems like JMP doesn't behave correctly with Value Labels when "." is one of the values which is being labeled. I'll report this as a bug when I have time
Names Default To Here(1);
dt = New Table("Untitled",
Add Rows(4),
New Column("Column 1",
Character,
"Nominal",
Value Labels({"." = "MISSING", "A" = "AAA", "B" = "BBB", "C" = "CCC"}),
Use Value Labels(1),
Set Selected,
Set Values({"A", "B", ".", "C"})
)
);- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Import x and y values from the text file to jmp table and create wafer map
Thanks Jarmo.
- « Previous
- Next »
Recommended Articles
- © 2026 JMP Statistical Discovery LLC. All Rights Reserved.
- Terms of Use
- Privacy Statement
- Contact Us