- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Discussions
Solve problems, and share tips and tricks with other JMP users.- JMP User Community
- :
- Discussions
- :
- How to analyze factorial CRD with subsamplings in JMP?
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
How to analyze factorial CRD with subsamplings in JMP?
I've been trying to analyze this design (one shot!) for a long time. Still could not. As I've refer more than 3 months, I see this is only possible in GenStat S/W.
My experiment is CRD: Two Factor Factorial Experiment, Fixed Effects.
Factor 1: Levels: a,b (2) Factor 2: Levels: p,q,r (3) for each experimental units, there are four subsamples (Observational units) I want to add subsampling to this model and do the analysis.
If possible could you please help me with JMP and if possible R codes as well for Two-way design (no blocking) with subsamples.
This is the example I got from GenStat S/W Page 48 (Two-way design (no blocking) with subsamples)
Also same example solved by SAS, in the book Contemporary Statistical Models for the Plant and Soil Sciences
Accepted Solutions
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: How to analyze factorial CRD with subsamplings in JMP?
*Note: This was my original reply, which at one point disappeared. I responded again before this reply reappeared.
Hi Kynda,
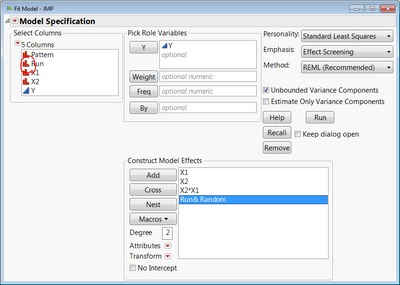
You would need a column to identify the run that produced each set of 4 repeat measures. Make sure that column has a nominal modeling type. In Fit Model, specify your model as usual, but add that column identifying each run and make it a random effect (select the Run effect in Construct Model Effects, and click Attributes > Random).
This is all assuming you have replicates in your design (like the stem length experiment has pots 1, 2 & 3 for each combination). If you don't replicate any of those factor level combinations in the experiment, then you're not going to be able to estimate the variance components for the Run blocking factor since it will be confounded with the X1*X2 interaction. JMP will basically ignore the Run random effect and analyze it as if you have 3 replicates (4 of each) rather than repeat measures which would be highly misleading. If you forego the interaction and just model main effects, you will be able to get the variance component for Run, but you will still be contending with how much of that variance component is really just X1*X2 interaction effects.
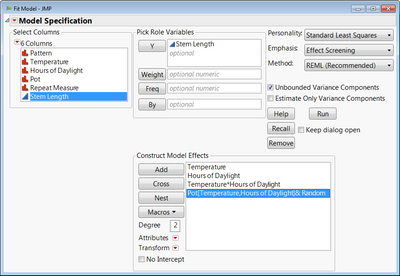
Here is an example with the stem length experiment from your first reference. I had to nest the Pot random effect into Hours of Daylight and Temperature so that Pot 1 for a given factor level combination is not treated as if it is the same block as another Pot 1 for a different factor level combination.
To do the nesting you would, just highlight the Pot effect, and then select Temperature and Hours of Daylight from the list of columns, and click "Nest". I'm attaching a data table with the analysis saved in a script.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: How to analyze factorial CRD with subsamplings in JMP?
*Note: This was my original reply, which at one point disappeared. I responded again before this reply reappeared.
Hi Kynda,
You would need a column to identify the run that produced each set of 4 repeat measures. Make sure that column has a nominal modeling type. In Fit Model, specify your model as usual, but add that column identifying each run and make it a random effect (select the Run effect in Construct Model Effects, and click Attributes > Random).
This is all assuming you have replicates in your design (like the stem length experiment has pots 1, 2 & 3 for each combination). If you don't replicate any of those factor level combinations in the experiment, then you're not going to be able to estimate the variance components for the Run blocking factor since it will be confounded with the X1*X2 interaction. JMP will basically ignore the Run random effect and analyze it as if you have 3 replicates (4 of each) rather than repeat measures which would be highly misleading. If you forego the interaction and just model main effects, you will be able to get the variance component for Run, but you will still be contending with how much of that variance component is really just X1*X2 interaction effects.
Here is an example with the stem length experiment from your first reference. I had to nest the Pot random effect into Hours of Daylight and Temperature so that Pot 1 for a given factor level combination is not treated as if it is the same block as another Pot 1 for a different factor level combination.
To do the nesting you would, just highlight the Pot effect, and then select Temperature and Hours of Daylight from the list of columns, and click "Nest". I'm attaching a data table with the analysis saved in a script.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: How to analyze factorial CRD with subsamplings in JMP?
I'm not sure why, but my earlier reply was deleted...
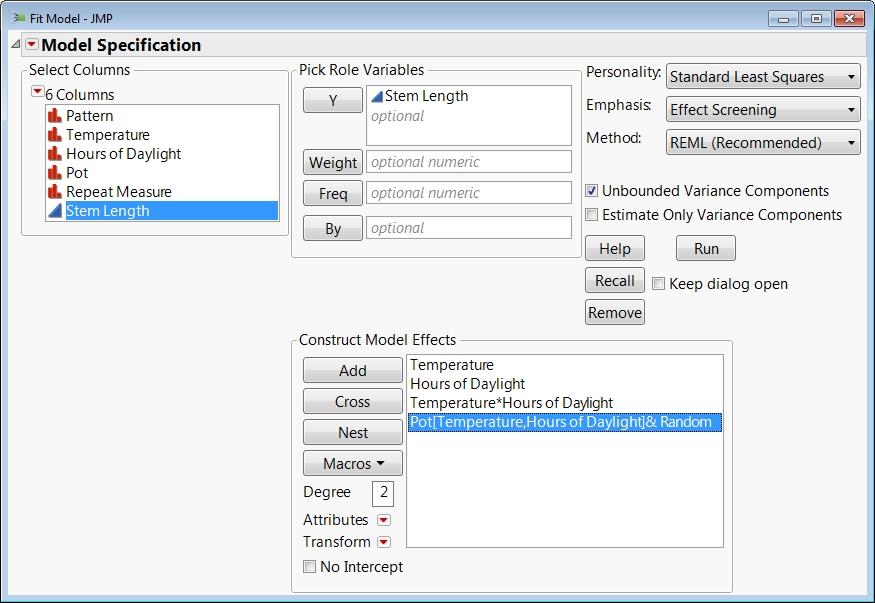
You basically need a column that groups the 4 repeat measures together. In the stem length example from your first reference, it would be the Pot (although this needs to be nested in the other 2 factors so that Pot 1 for high temperature, 8 hours of sunlight is not treated as the same block as Pot 1 for low temperature at 16 hours of sunlight). It is important you make sure that this grouping column is a nominal modeling type, and it's going to act as a random block in our model.
In Fit Model, you would specify your factorial model as usual, but you then add the grouping column for your repeat measures to the list of model effects. Then select the grouping column in the list of effects and click Attributes > Random to make it a random effect.
As previously stated, in the stem length experiment case we need to nest this effect within Temperature and Hours of Sunlight. You do this by selecting Pot in the Construct Model Effects Box and then select Temperature and Hours of Sunlight in the list of columns and click "Nest." This would not be necessary if each group had a unique name in the column. The final result should look like this:
I'm attaching the data table with the analysis saved in a script.
Recommended Articles
- © 2026 JMP Statistical Discovery LLC. All Rights Reserved.
- Terms of Use
- Privacy Statement
- Contact Us