- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Discussions
Solve problems, and share tips and tricks with other JMP users.- JMP User Community
- :
- Discussions
- :
- Re: Hierarchical clustering: JMP & R disagree
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Hierarchical clustering: JMP & R disagree
When I cluster the same exact dataset in JMP and in R, using Ward's method, I get results that are non-identical. R provides two options (ward.D and ward.D2), wherein the latter squares the values in the distance matrix before clustering them. According to Murtagh & Legendre (2014), that's what JMP does too. But even when I specify method="ward.D2" in R, the output I get is non-trivially different: roughly 15% of the observations are mismatched with respect to what I get from JMP.
So, Question 1: Am I wrong to expect that the same method should produce the same results in HCA? I wouldn't expect that in k-means clustering (without setting a seed, at least), but for HCA the results are totally stable *within* a given package, so I assumed they should also be the same *between* packages.
Question 2: If they *should* be the same... why aren't they?
- Possibility A: there's a difference in how the distance matrix is calculated
- Possibility B: there's a difference in how Ward's method is implemented
- Possibility C: the cutree() function in R does something different than the "Select number of clusters" function in JMP
- Possibility X... ???
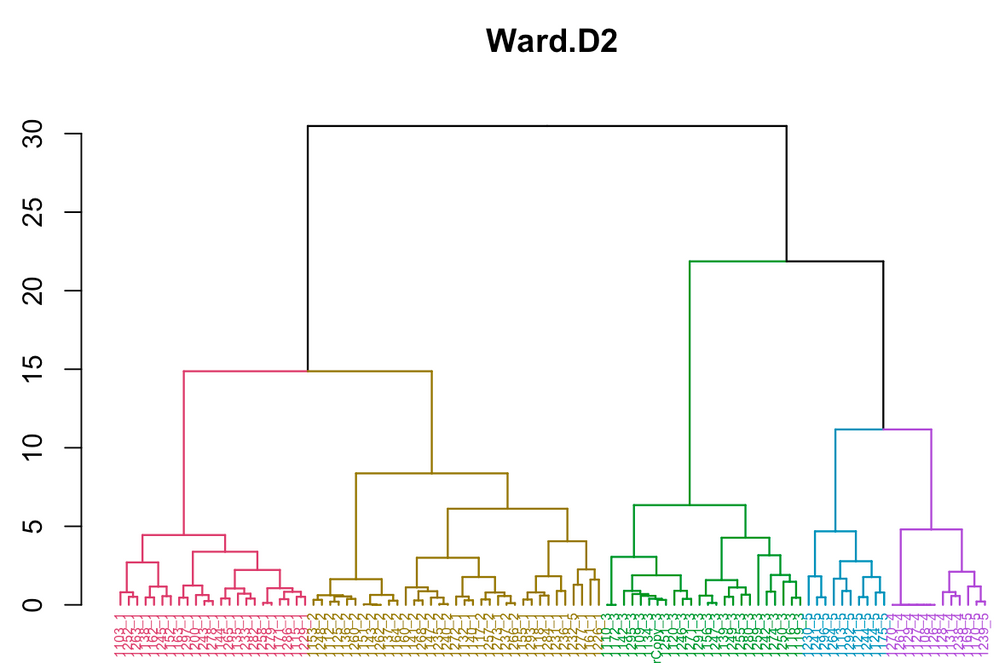
I've uploaded the dendrogram from R (apologies if that's poor netiquette!); you can see the problem I'm talking about by taking a close look at the labels along the x-axis. Those labels all end in a digit from 1-5, which represents the number of the cluster they were assigned to in JMP. As you can see, the green group ("3") was the only one where there was a perfect 1:1 match between JMP and R.
Grateful for any guidance, -Matt
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Hierarchical clustering: JMP & R disagree
Hi matthal_resear,
Comparing different packages is always kind of a hot topic. As I cannot take a look at the source codes in JMP I cannot check if there is a difference in any of the calculations you mentioned in point 2 between JMP and R. They both refer to the Murtagh & Legendre (2014), however this does not mean that the algorithm is implemented in the complete similar fassion. Especially before and after the core of the algorithm might be differences. Also in potential subroutines. So again, as there is no way to check the complete code one will not be able to answer this question. you may try to send your question to JMP's Technical Support, providing the detailed information about the R-Package you are using as well the concrete routine. Sometimes there are more than one R function available. Then they might have a chance to check for differences, not saying they will find something.
Just another thought. Are the results within the packages stable for HCA? Or are they slightly different? This is also an important information. Last I would take a look at the constellation plot to identify differences there as well.
Just my2cents
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Hierarchical clustering: JMP & R disagree
This thread might also help with the distance matrix:
https://community.jmp.com/t5/Discussions/which-distance-does-JMP-use-in-clustering/m-p/275384#M53438
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Hierarchical clustering: JMP & R disagree
Thanks for passing that along; it made me realize that it will be worth actually exporting the distance matrix from JMP and comparing those values to the distance matrix I get in R! That will be one way to narrow down where the discrepancy is coming from.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Hierarchical clustering: JMP & R disagree
Thanks for the suggestion; it's good to know that sending the code to the tech team is an option if I can't resolve this another way.
To answer your other question, yes: the HCA results within each package are completely stable.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Hierarchical clustering: JMP & R disagree
Is it possible that the data are being standardized in one package and not the other? The default in JMP is "Standardize Data".
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Hierarchical clustering: JMP & R disagree
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Hierarchical clustering: JMP & R disagree
Hmmm... I just did a comparison on three different data sets in JMP and R (without using "standardize data" or "scale()") and found the classifications to match exactly in each when using the same number of clusters. The only methods I specified in R were to use 'euclidean' for the "dist" function and 'ward.D2' for the "hclust" function. Are you specifying anything else in your R and/or JMP workflows? How messy are your data (missing... outliers...)? Just offering some food for thought...
Recommended Articles
- © 2026 JMP Statistical Discovery LLC. All Rights Reserved.
- Terms of Use
- Privacy Statement
- Contact Us