- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Discussions
Solve problems, and share tips and tricks with other JMP users.- JMP User Community
- :
- Discussions
- :
- Re: FCS Files: JMP opens them, but where does the header information go?
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
FCS Files: JMP opens them, but where does the header information go?
I am helping a colleague with some work with FCS files. JMP opens the data easy enough, which is great, but I am looking to extract the header information, and I can't figure out an easy way - especially if I wanted to do this for a bunch of files.
I don't get any options like I do for CSV files, so the only solution I have found is to change the extension to .txt and open the file with text preferences. That works, but would seem troublesome if I need to do this to all files prior to opening.
1) is there a way to open a single FCS file to see the header w/o changing the extension
2) can you use a script and force-use text preferences even if the extension isn't a standard text extension.
Thanks!
Accepted Solutions
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: FCS Files: JMP opens them, but where does the header information go?
I took the code a bit further to unpick some of the fields in the header
fcsFile = "my-fcs.fcs";
str = loadtextfile(fcsFile);
// extract header
pat = "FCS" + PatArb()>>hdr + "$BEGINANALYSIS";
isMatch = PatMatch(str,pat);
if (!isMatch,
throw("oops")
);
// extract selected header info
system = ""; // computer system
pat = "$SYS" + "|" + PatArb()>>system + "|";
PatMatch(hdr,pat);
cytometer = ""; // type of cytometer
pat = "$CYT" + "|" + PatArb()>>cytometer + "|";
PatMatch(hdr,pat);
startTime = ""; // start time
pat = "$BTIM" + "|" + PatArb()>>startTime + "|";
PatMatch(hdr,pat);
endTime = ""; // end time
pat = "$ETIM" + "|" + PatArb()>>endTime + "|";
PatMatch(hdr,pat);
show(system,cytometer,startTime,endTime);For my test data file this gives
system = "Summit V5.2.0.7477 / Windows NT Version 5.1 / Intel Processor"; cytometer = "MoFlo XDP"; startTime = "12:34:07"; endTime = "12:38:15";
I used some field identifiers based on this documentation
https://www.bioconductor.org/packages/release/bioc/vignettes/flowCore/inst/doc/fcs3.html#3.1
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: FCS Files: JMP opens them, but where does the header information go?
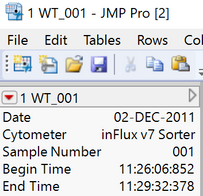
When JMP opens an FCS file it places the header information as table variables.
You can interrogate the table to get the list of names (dt<<getTableVariableNames) and you can retrieve the values (
dt<<getTableVariableNames).
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: FCS Files: JMP opens them, but where does the header information go?
@David_Burnham, That would be awesome! ... but I am not seeing that. I am on JMP 15.2.1 on Windows 10.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: FCS Files: JMP opens them, but where does the header information go?
One option you might want to try is using the function LoadTextFile, this will at least give you visibility to the header data (although can be slow because FCS files contain large blocks of binary data):
fcsFile = "c:\my-fcs-file.fcs";
str = loadtextfile(fcsFile);
// extract header
pat = "FCS" + PatArb()>>hdr + "$BEGINANALYSIS";
isMatch = PatMatch(str,pat);
if (isMatch,
show(hdr)
,
show("oops")
);
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: FCS Files: JMP opens them, but where does the header information go?
Maybe this will give me a hand understanding why...
Thanks!
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: FCS Files: JMP opens them, but where does the header information go?
I took the code a bit further to unpick some of the fields in the header
fcsFile = "my-fcs.fcs";
str = loadtextfile(fcsFile);
// extract header
pat = "FCS" + PatArb()>>hdr + "$BEGINANALYSIS";
isMatch = PatMatch(str,pat);
if (!isMatch,
throw("oops")
);
// extract selected header info
system = ""; // computer system
pat = "$SYS" + "|" + PatArb()>>system + "|";
PatMatch(hdr,pat);
cytometer = ""; // type of cytometer
pat = "$CYT" + "|" + PatArb()>>cytometer + "|";
PatMatch(hdr,pat);
startTime = ""; // start time
pat = "$BTIM" + "|" + PatArb()>>startTime + "|";
PatMatch(hdr,pat);
endTime = ""; // end time
pat = "$ETIM" + "|" + PatArb()>>endTime + "|";
PatMatch(hdr,pat);
show(system,cytometer,startTime,endTime);For my test data file this gives
system = "Summit V5.2.0.7477 / Windows NT Version 5.1 / Intel Processor"; cytometer = "MoFlo XDP"; startTime = "12:34:07"; endTime = "12:38:15";
I used some field identifiers based on this documentation
https://www.bioconductor.org/packages/release/bioc/vignettes/flowCore/inst/doc/fcs3.html#3.1
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: FCS Files: JMP opens them, but where does the header information go?
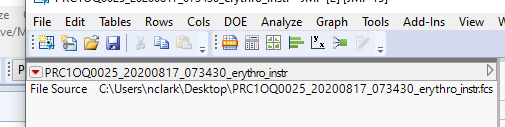
Thanks! I will need to touch base with support to see why this file doesn't have the header info in the table vars, but I can use the script you sent to at least dig into the files. Even with the overhead of loading an entire file, it beats the alternatives :)
-Nate
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: FCS Files: JMP opens them, but where does the header information go?
Yes I am curious why you are not seeing the header information. I would guess either it is the version of the FCS files (I was testing with FCS3.0), or JMP is selective in the header information that it looks for.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: FCS Files: JMP opens them, but where does the header information go?
I'm pretty sure JMP has a list of standard keys it recognizes and may ignore other (vendor specific?) keys.
There is also information in each column's properties, maybe for interpreting ranges.
@Anonymous @briancorcoran
Recommended Articles
- © 2026 JMP Statistical Discovery LLC. All Rights Reserved.
- Terms of Use
- Privacy Statement
- Contact Us