- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Discussions
Solve problems, and share tips and tricks with other JMP users.- JMP User Community
- :
- Discussions
- :
- Re: Disallowed Combinations not working
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Disallowed Combinations not working
Hi,
I am trying to make disallowed combinations in my custom design.
I have a factor with two categorical levels: No ChCl and ChCl and a factor with three ChCl molar ratio (categorical): 0, 1 and 2
I want to make a Disallowed Combinations where 'No ChCl' and a molar ratio of 0 cannot be made.
ChCl== "No ChCl" & Molar ratio ChCl== '0'
But the design still makes a combination of No ChCl and 0. What am I doing wrong?
best regards, Freja
Accepted Solutions
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Disallowed Combinations not working
Hi @frejam,
I would recommend setting the factor ML HBA as a discrete numeric with three levels instead of a 3-levels categorical factor, as the ordering between levels is taken into consideration with the discrete numeric factor type, and it does suit better your experimental design from what I see. It also facilitates the creation of the disallowed combinations constraint.
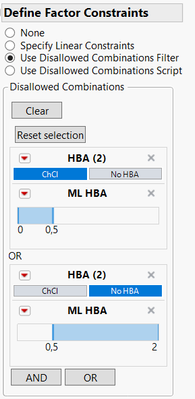
If I set up the disallowed combinations like these :
Or in a script version :
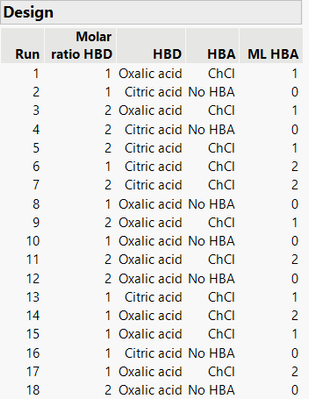
HBA == "ChCl" & ML HBA <= 0.5 | HBA == "No HBA" & ML HBA >= 0.5Then the generated design seems to respect the constraint you want:
Here is the script to generate the design (I assumed a model with main effects and 2-factors interactions) :
DOE(
Custom Design,
{Add Response( Maximize, "Y", ., ., . ),
Add Factor( Continuous, 1, 2, "Molar ratio HBD", 0 ),
Add Factor( Categorical, {"Oxalic acid", "Citric acid"}, "HBD", 0 ),
Add Factor( Categorical, {"ChCl", "No HBA"}, "HBA", 0 ),
Add Factor( Discrete Numeric, {0, 1, 2}, "ML HBA", 0 ),
Set Random Seed( 2109323028 ), Number of Starts( 3867 ), Add Term( {1, 0} ),
Add Term( {1, 1} ), Add Term( {2, 1} ), Add Term( {3, 1} ), Add Term( {4, 1} ),
Add Potential Term( {4, 2} ), Add Term( {1, 1}, {2, 1} ),
Add Term( {1, 1}, {3, 1} ), Add Term( {1, 1}, {4, 1} ),
Add Term( {2, 1}, {3, 1} ), Add Term( {2, 1}, {4, 1} ),
Add Term( {3, 1}, {4, 1} ), Set Sample Size( 18 ),
Disallowed Combinations(
HBA == "ChCl" & ML HBA <= 0.5 | HBA == "No HBA" & ML HBA >= 0.5
), Simulate Responses( 0 ), Save X Matrix( 0 ), Make Design}
)And you can find the datatable attached.
The analysis should be done carefully, as you might have a nested effect of "ML HBA" inside "HBA" : the level 0 for factor "ML HBA" is only accessible for the level No HBA of factor "HBA".
Hope this answer will help you,
"It is not unusual for a well-designed experiment to analyze itself" (Box, Hunter and Hunter)
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Disallowed Combinations not working
Hi @frejam,
Looking at the new post Design of Experiment - Optional Mixture Additives, I wonder if your situation is not similar (since factors HBA and ML HBA are related ?) and the required design may not be more simple than what I suggested ?
Would it make sense for your experimental setup to consider creating a design with 3 factors :
- Molar ratio HBD : continuous factor, range 1-2
- HBD : 2-levels categorical factor, oxalic acid or citric acid
- ML HBA : continuous factor, range 0-2
This setup enables to avoid nested factors, simplify the analysis and covers a broader and continuous experimental space. The situation with "No HBA" would be represented when ML HBA has the value 0.
To have three levels for the factors ML HBA, you can enter the quadratic effect of this factor in the Model panel :
The design obtained with the default number of runs look like this :
Here is the script to generate the design :
DOE(
Custom Design,
{Add Response( Maximize, "Y", ., ., . ),
Add Factor( Continuous, 1, 2, "Molar ratio HBD", 0 ),
Add Factor( Categorical, {"Oxalic acid", "Citric acid"}, "HBD", 0 ),
Add Factor( Continuous, 0, 2, "ML HBA", 0 ), Set Random Seed( 1591090212 ),
Number of Starts( 109881 ), Add Term( {1, 0} ), Add Term( {1, 1} ),
Add Term( {2, 1} ), Add Term( {3, 1} ), Add Term( {1, 1}, {2, 1} ),
Add Term( {1, 1}, {3, 1} ), Add Term( {2, 1}, {3, 1} ), Add Term( {3, 2} ),
Set Sample Size( 12 ), Simulate Responses( 0 ), Save X Matrix( 0 ), Make Design}
)And design datatable is attached. Feel free to adjust the sample size and assumed model to your needs.
I hope this suggestion can help you and simplify your design and project,
"It is not unusual for a well-designed experiment to analyze itself" (Box, Hunter and Hunter)
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Disallowed Combinations not working
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Disallowed Combinations not working
Hi Georg,
I don't see that it works in your example either?
So here I applied the constrain, but I still combination where molar ratio (ML HBA) is 0 and HBA is ChCl.
What I want is that if HBA = ChCl then ML HBA should be >0, or if HBA = No ChCl then ML HBA = 0. Does that make sense? I attached my DoE table (I hope)
Best regards Freja
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Disallowed Combinations not working
Hi @frejam,
I would recommend setting the factor ML HBA as a discrete numeric with three levels instead of a 3-levels categorical factor, as the ordering between levels is taken into consideration with the discrete numeric factor type, and it does suit better your experimental design from what I see. It also facilitates the creation of the disallowed combinations constraint.
If I set up the disallowed combinations like these :
Or in a script version :
HBA == "ChCl" & ML HBA <= 0.5 | HBA == "No HBA" & ML HBA >= 0.5Then the generated design seems to respect the constraint you want:
Here is the script to generate the design (I assumed a model with main effects and 2-factors interactions) :
DOE(
Custom Design,
{Add Response( Maximize, "Y", ., ., . ),
Add Factor( Continuous, 1, 2, "Molar ratio HBD", 0 ),
Add Factor( Categorical, {"Oxalic acid", "Citric acid"}, "HBD", 0 ),
Add Factor( Categorical, {"ChCl", "No HBA"}, "HBA", 0 ),
Add Factor( Discrete Numeric, {0, 1, 2}, "ML HBA", 0 ),
Set Random Seed( 2109323028 ), Number of Starts( 3867 ), Add Term( {1, 0} ),
Add Term( {1, 1} ), Add Term( {2, 1} ), Add Term( {3, 1} ), Add Term( {4, 1} ),
Add Potential Term( {4, 2} ), Add Term( {1, 1}, {2, 1} ),
Add Term( {1, 1}, {3, 1} ), Add Term( {1, 1}, {4, 1} ),
Add Term( {2, 1}, {3, 1} ), Add Term( {2, 1}, {4, 1} ),
Add Term( {3, 1}, {4, 1} ), Set Sample Size( 18 ),
Disallowed Combinations(
HBA == "ChCl" & ML HBA <= 0.5 | HBA == "No HBA" & ML HBA >= 0.5
), Simulate Responses( 0 ), Save X Matrix( 0 ), Make Design}
)And you can find the datatable attached.
The analysis should be done carefully, as you might have a nested effect of "ML HBA" inside "HBA" : the level 0 for factor "ML HBA" is only accessible for the level No HBA of factor "HBA".
Hope this answer will help you,
"It is not unusual for a well-designed experiment to analyze itself" (Box, Hunter and Hunter)
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Disallowed Combinations not working
Hi Victor,
Thank you so much for helping out. This was exactly the solution I needed :)
Best regards, Freja
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Disallowed Combinations not working
Hi again,
Can you expand on what you mean by this sentence?:
"The analysis should be done carefully, as you might have a nested effect of "ML HBA" inside "HBA" : the level 0 for factor "ML HBA" is only accessible for the level No HBA of factor "HBA".
I can see that I get a very low score for HBA and ML HBA (<0.4) in my power analysis when I put on the constrain. Is this effect you are talking about above and any suggestions to improve this?
Sorry for all my questions - I am new in DoE design.
Best regards, Freja
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Disallowed Combinations not working
Hi @frejam,
What I mean with :
"The analysis should be done carefully, as you might have a nested effect of "ML HBA" inside "HBA" : the level 0 for factor "ML HBA" is only accessible for the level No HBA of factor "HBA".
The factors "HBA" and "ML HBA" are not independent, since the levels of "ML HBA" depend on the choice of the level for "HBA". This situation reduce the effective detection of these effects, as they are not independant (you can see in your power analysis). There might be no solution to this, increasing the sample size might help but won't solve entirely the problem.
For example, if HBA is set to "No HBA", then the only possible value for "ML HBA" is 0. This dependance creates a nesting effect : "ML HBA" is nested within factor "HBA". Or if you want to see this situation and your experimental design more easily and graphically :
Let's see how this situation might impact your regression model through a simulated example. I created a response formula Y with the following equation :
If we simply click on script "Model" with the assumed model from the generated design (or use script "Fit Least Squares (normal)"), with all main effects and 2-factors interactions estimated independently, we encounter some errors due to the non-independance between ML HBA and HBA:
- There is a "Singularity details" panel added, showing linear dependancies among our predictors :
- Looking at the "Effects Summary" panel, not all effects can be estimated (even if we generated a design able to fit these terms/effects):
- Looking at the "Parameter Estimates" panel, we see that main effects HBA and ML HBA are noted as biased (and estimated with a relatively large standard error), which again is an indication that they couldn't be estimated independantly :
All these informations show indication that our regression model may not be appropriate in the context of nested factors. In order to solve this problem, you can manually specify the effects contained in your regression model through the Fit Model platform to include a nesting effect of ML HBA within HBA :
To create nesting between factors, you can check this section of JMP Help : Example of a Two-Factor Nested Random Effects Model (jmp.com)
Since not all factors are crossed, it's only possible to estimate 2-factors interactions for factors that are not nested. In this case, you could add interactions between "HBD" and "Molar ratio HBD", "HBD" and "HBA", and "Molar ratio HBD" and "HBA".
Running this model (use script "Fit Least Squares (nested)"), you can see there is no singularity anymore, all effects can be estimated, an no terms coefficients are biased. You can also see that coefficients for the terms are close to the one specified in the formula seen before, so the full model seems to be quite appropriate :
Of course, depending on your performance metric (RMSE, R² and R² adjusted, statistical significance, information criterion like AICc and BIC, ...), you could further refine the model. But you should start first with your complete model assumed from your experimental design.
I join the datatable with all scripts and formula used for this example.
I hope this example will help you understand better the challenge of analyzing nested factors in DoE.
Best,
"It is not unusual for a well-designed experiment to analyze itself" (Box, Hunter and Hunter)
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Disallowed Combinations not working
Thank you so much for your explanation including examples, Victor. It really helped in understanding what nested effect is and how to overcome it. I will try to apply it to my design.
Best regards, Freja
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Disallowed Combinations not working
Hi @frejam,
Looking at the new post Design of Experiment - Optional Mixture Additives, I wonder if your situation is not similar (since factors HBA and ML HBA are related ?) and the required design may not be more simple than what I suggested ?
Would it make sense for your experimental setup to consider creating a design with 3 factors :
- Molar ratio HBD : continuous factor, range 1-2
- HBD : 2-levels categorical factor, oxalic acid or citric acid
- ML HBA : continuous factor, range 0-2
This setup enables to avoid nested factors, simplify the analysis and covers a broader and continuous experimental space. The situation with "No HBA" would be represented when ML HBA has the value 0.
To have three levels for the factors ML HBA, you can enter the quadratic effect of this factor in the Model panel :
The design obtained with the default number of runs look like this :
Here is the script to generate the design :
DOE(
Custom Design,
{Add Response( Maximize, "Y", ., ., . ),
Add Factor( Continuous, 1, 2, "Molar ratio HBD", 0 ),
Add Factor( Categorical, {"Oxalic acid", "Citric acid"}, "HBD", 0 ),
Add Factor( Continuous, 0, 2, "ML HBA", 0 ), Set Random Seed( 1591090212 ),
Number of Starts( 109881 ), Add Term( {1, 0} ), Add Term( {1, 1} ),
Add Term( {2, 1} ), Add Term( {3, 1} ), Add Term( {1, 1}, {2, 1} ),
Add Term( {1, 1}, {3, 1} ), Add Term( {2, 1}, {3, 1} ), Add Term( {3, 2} ),
Set Sample Size( 12 ), Simulate Responses( 0 ), Save X Matrix( 0 ), Make Design}
)And design datatable is attached. Feel free to adjust the sample size and assumed model to your needs.
I hope this suggestion can help you and simplify your design and project,
"It is not unusual for a well-designed experiment to analyze itself" (Box, Hunter and Hunter)
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Disallowed Combinations not working
Hi Victor,
Thank you for following up on this. Changing the Molar ratio to continuous as you suggested improves the power of my design. So I will definitely use this approach. Thank you so much.
Best regards, Freja
Recommended Articles
- © 2026 JMP Statistical Discovery LLC. All Rights Reserved.
- Terms of Use
- Privacy Statement
- Contact Us