- Subscribe to RSS Feed
- Mark as New
- Mark as Read
- Bookmark
- Subscribe
- Printer Friendly Page
- Report Inappropriate Content
JMP Add-Ins
Download and share JMP add-ins- JMP User Community
- :
- File Exchange
- :
- JMP Add-Ins
- :
- Bioassay
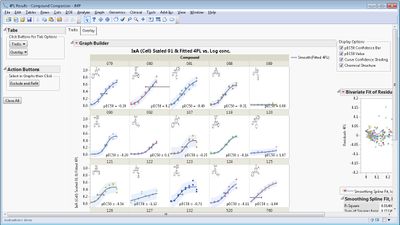
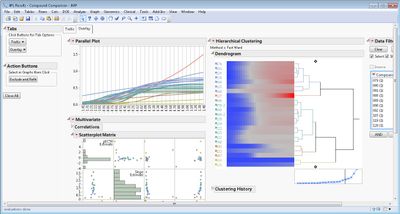
This add-in helps you analyze and compare multiple bioassay curves fit with a 4-Parameter or 5-Parameter Logistic (4PL/5PL) model. It calls the Nonlinear platform behind the scenes to fit the models to each curve and then creates a variety of graphical and tabular results.
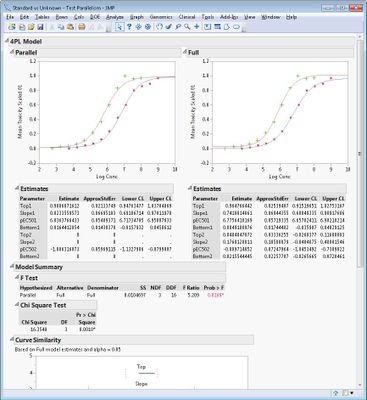
The add-in contains two routines, Fit Curves and Test Parallelism. Details about them are available by clicking Add-Ins > Bioassay > Help after installing the add-in.
To install the add-in, download "Bioassay.jmpaddin", drag it onto an open JMP window, then click "Install". The add-in includes the three example data sets, "Compound Comparison.jmp", "Chemical Structures.jmp", and "Standard vs Unknown.jmp". These are used for illustration in the help documentation.
Some example screen shots are below.
This is a very useful add-in. How I can change it from EC50 to EC80
- Could you please give me some information of what the "chisquare test" in the Bioassay Add-in represents? (formula)
Is this chisquare value presented in JMP for the goodness of fit? in other words, the sum of the weighted residuals of the constrained fit? or does it take to account the unconstrained, full model. We tried to calculate the sum of the weighted residuals of the constrained fit and we never get the number presented.
Hi could someone either provide or route me to the download section to retrieve the three mentoned example data sets, "Compound Comparison.jmp", "Chemical Structures.jmp", and "Standard vs Unknown.jmp"? Was there any progress in determining EC80 (see previous post by "Rao, 09-27-2018". Thank you!
Best, Nico
Hi @nicoja The data sets are included with the add-in; look under the Example Data submenu. EC80 can be derived from the logistic parameter estimates using a formula like the following: EC80 = EC50 * (0.8/(1-0.8))^(1/Slope)
- Mark as Read
- Mark as New
- Bookmark
- Get Direct Link
- Report Inappropriate Content
Dear @russ_wolfinger,
Do you mind to share with me the statistic equation or formula of the Lower CL and Upper CL, when getting relative potency of EC50?
I used Statlia software with same set of raw data, and got the same coef a, b, c, d of the 4 PL curve and same relative potency, but the lower and upper CL for EC50 were different. I just want to figure out why those are different.
Thanks,
Emily
Hi Emily @EnsembleScore81 ,
The definitive source is the JSL code itself. Click View > Add-Ins, select Bioassay, then click the link to the Home Folder. In that folder is a file Analysis Functions.jsl, and look starting at line 438.
In case you want to try, you can also now compute Relative Potency directly in the JMP Fit Curve platform. Here are some relevant links:
https://community.jmp.com/t5/JMPer-Cable/How-to-perform-Bioassay-analysis-with-JMP/ba-p/584762
https://community.jmp.com/t5/Discussions/Relative-Potancy-95-CI/td-p/326815
Kindly,
Russ
Recommended Articles
- © 2026 JMP Statistical Discovery LLC. All Rights Reserved.
- Terms of Use
- Privacy Statement
- Contact Us