- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Discussions
Solve problems, and share tips and tricks with other JMP users.- JMP User Community
- :
- Discussions
- :
- Re: A call to JMP Scientists
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Setting up 96 and 384 well plate designs with replicates
Hey JMP scientists,
I’m working on some cell based assay development projects where I’m using DOEs to try to optimize the parameters I can control in the experiments.
Setting up the layouts for 96 and 384 well plates to get a reasonable level of replication and avoid edge effects is kind of a pain. Are there some good strategies that don’t involve randomizing the entire plate with a robot? (although I have a liquid handler for certain variables which are more amenable to randomization) What kinds of designs and what terms (e.g. interactions, quadratic, cubic..) are you putting into the designs? How do you handle 'replicates' in the DOE custom design generator. I am curious to get as much feedback as possible. I understand the power and utility of DOE, I am a convert, but I just need help finally getting this effort off the ground and utilizing it in our day to day assay development and experimental design.
Thank you!
Accepted Solutions
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: A call to JMP Scientists
Building 96 well experiments.
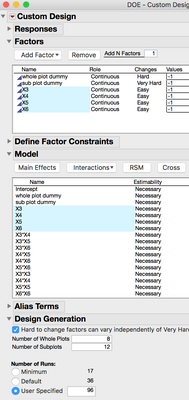
Take a look at this DOE construction method. There are 4 continuous factors, and 2 additional continuous factors to use as dummy variables for the hard ad very hard to change settings. (dummy variables to construct the whole and subplots.) Factors X3, X4, X5, and X6 are the experimental factos, the design includes interaction and quadratic model terms for each of them. Factors X1 and
X2 are main effects that are set as hard and very hard to change.
(Translated from Stat Speak: If you've got a robot that can pipet a random plate layout, it would be convenient if your DOE table also had plate locations in it too. In this example, I'm going to force JMP to give me plate positions which will be labeled "Subplot" and "Whole Plot." In order to make this work, the first two factors/variables/inputs/X's I add are just going to be dummys. Make the first dummy X hard to change, and make the second X very hard to change. From here add your other variables like drug, inhibitor, buffer, co-factor, nutrient, carbon source or whatever you're interested in studying. Then like normal, add the terms, like interactions and quadratics to the model. Be very careful not to include the dummy X's in the interactions and quadratic terms. All we need are the main effects.
This last step is critical to making this work, don't skip this. There is an option, "Hard to change factors can vary independently of very hard to change factors." This MUST be checked. Take a look at the script and you will notice this: "Make Strip Plot Design", in the arugument. Turning on this option makes that bit occur in the script. (Stats Speak: JMP can make strip plot designs with restrictions on randomization for whole and subplots that are independed of each other.) The next part is important too. Set the number of Whole Plots and Sub Plots, and then the number of runs. One of the plots should be 8 and the other should be 12, and the number of runs is 64. In the past I would have only 8 columns reserved for the experiment, and then use the last three for controls and a standard curves. In this case, Whole Plots and Sub Plots are both 8 and runs = 64. After you make the design table it shouldn't be too hard to pass the design to your robot for pipetting.)
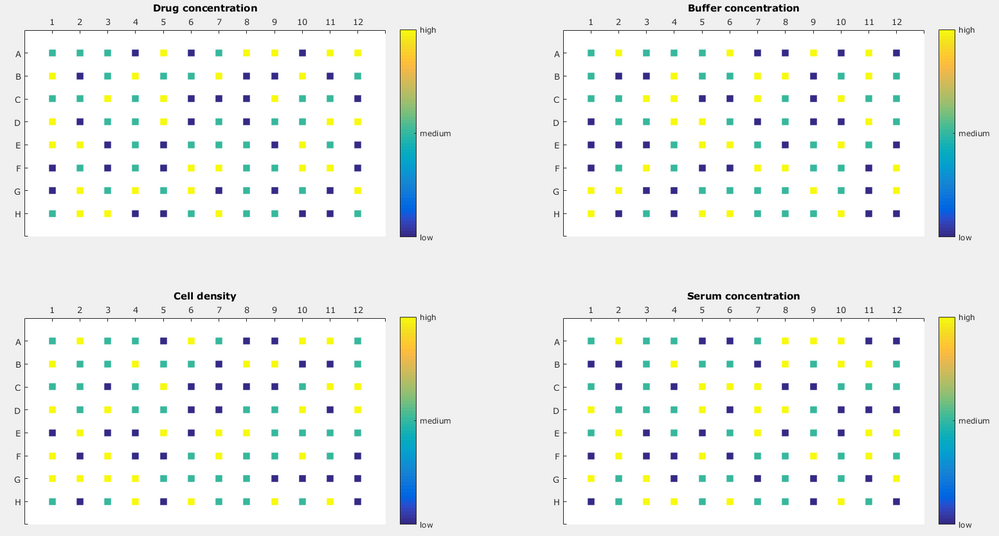
In the design that is generated, JMP provides the 8 whole and 12 subplots in two columns that can be used as the location of each of the 96 wells. In the figure below, X3, X4, X5 and X6 have no restrictions on randomization, and a different pattern is evident for each variable, represented as four the four grids.
After the experiment is complete, it will be important to remember not to include the two dummy variables in the model. By default these will be in the model script.
Design Script
DOE(
Custom Design,
{Add Response( Maximize, "Y", ., ., . ),
Add Factor( Continuous, -1, 1, "whole plot dummy", 1 ),
Add Factor( Continuous, -1, 1, "sub plot dummy", 2 ),
Add Factor( Continuous, -1, 1, "X3", 0 ),
Add Factor( Continuous, -1, 1, "X4", 0 ),
Add Factor( Continuous, -1, 1, "X5", 0 ),
Add Factor( Continuous, -1, 1, "X6", 0 ), Set Random Seed( 42 ),
Number of Starts( 10 ), Add Term( {1, 0} ), Add Term( {1, 1} ),
Add Term( {2, 1} ), Add Term( {3, 1} ), Add Term( {4, 1} ), Add Term( {5, 1} ),
Add Term( {6, 1} ), Add Term( {3, 1}, {4, 1} ), Add Term( {3, 1}, {5, 1} ),
Add Term( {3, 1}, {6, 1} ), Add Term( {4, 1}, {5, 1} ),
Add Term( {4, 1}, {6, 1} ), Add Term( {5, 1}, {6, 1} ), Add Term( {3, 2} ),
Add Term( {4, 2} ), Add Term( {5, 2} ), Add Term( {6, 2} ),
Make Strip Plot Design, Set N Whole Plots( 8 ), Set N Subplots( 12 ),
Set Sample Size( 96 ), Optimality Criterion( 2 ), Make Design}
);
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: A call to JMP Scientists
I've been lurking on this question because I'm sure there are likely some stong opinions on this topic. Here's one way to set up a 96 well experiment.
This design does some things that I think are useful.
In the experiment there are continuous factors:
1. Drug, an inhibitor I'm interested in studying
2. Stim, the thing stimulates a response the drug is supposed to inhibit
3. Serum level, grows cells better but increases background of the response
and some categorical factors:
1. Media type
2. Cell line, different cell line for each plate
To make the plate setup reasonable I want to place some restrictions on randomization. If I had lots of time I could program the pipetting robot to fully randomize the entire plate, but if just restrict randomization I can keep concentrations down columns and across rows the same, and plating gets a lot more simple.
First of all, for the scope of this, I essentially have 384 wells to work with, 96 per plate, and 4 plates (one for each cell line.) For the factors I'm working with, I can easily look at 2nd order interactions and quadratic effects for the continuous terms. When I set up the plates I have 12 columns and 32 rows (8 rows for each of the 4 plates.)
In the DOE dialog, I'm going to restrict the randomzation for rows and columns. This means making some factors "hard" to change and some "very hard.", which makes this a split strip plot experiment. Whole plots correspond to hard, and Subplots correspond to very hard to change factors. So if I have 12 subplots, and 32 (8x4) whole plots, then the hard to change things (in this case Drug and Serum) will be constant down the columns and the each indifidual row will be the same across the plate. Let me explain this a little more. Each column has a different concnetration, the concentration is the same down the entire column. (Super handy if you're multichannel pipetting your own plates.)
The last thing to consider is the plate/cell line. What I really need is a three way split plot, so cell line/plate would be "Super Hard to Change", but that's not an option, and It might be better if there was a different subplot pattern for the columns for each plate, but this is pretty good. (realistically, just making 8 whole plots and 12 subplots and using the same plate layout for each of the four cell lines is probably good enough, plus it reduces opportunities for error in the plate layout.)
DOE(
Custom Design,
{Add Response( Maximize, "Y", ., ., . ),
Add Factor( Continuous, 2, 25, "Drug", 1 ),
Add Factor( Continuous, 0.02, 0.1, "Serum", 1 ),
Add Factor( Categorical, {"IMDM", "RPMI", "DMEM"}, "Media", 2 ),
Add Factor( Continuous, 5, 50, "Stim", 2 ),
Add Factor( Categorical, {"L1", "L2", "L3", "L4"}, "Plate Number", 2 ),
Set Random Seed( 550796982 ), Number of Starts( 1 ), Add Term( {1, 0} ),
Add Term( {1, 1} ), Add Term( {2, 1} ), Add Term( {3, 1} ), Add Term( {4, 1} ),
Add Term( {5, 1} ), Add Term( {1, 2} ), Add Term( {1, 1}, {2, 1} ),
Add Term( {2, 2} ), Add Term( {1, 1}, {3, 1} ), Add Term( {2, 1}, {3, 1} ),
Add Term( {1, 1}, {4, 1} ), Add Term( {2, 1}, {4, 1} ),
Add Term( {3, 1}, {4, 1} ), Add Term( {4, 2} ), Add Term( {1, 1}, {5, 1} ),
Add Term( {2, 1}, {5, 1} ), Add Term( {3, 1}, {5, 1} ),
Add Term( {4, 1}, {5, 1} ), Add Term( {1, 3} ), Add Term( {1, 4} ),
Add Term( {4, 3} ), Make Strip Plot Design, Set N Whole Plots( 32 ),
Set N Subplots( 12 ), Set Sample Size( 384 ), Optimality Criterion( 2 )
);- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: A call to JMP Scientists
I think that this matter of designing a sophisticated experiment with restrictions on the randomization is important and, therefore, this discussion could help a lot of users in a similar situation. It is a great example of what we hoped this discussion area in the JMP Community could be for all of us. I am bewildered that there has been no activity for a week. I doubt that it is because the last reply answered all of the remaining questions.
I hope that the discussion picks up again. Here are some points that I hope provoke more replies:
- How is this case like your own if you develop assays? How is it different?
- Do you have the same restrictions on randomizing the runs (wells)? More? Less? Just different?
- The restrictions on the randomization clearly impact how practical the design is to run. Is it just as clear how the restriction impacts the analysis? What might happen to my interpretation of the regression model if the restriction is ignored in the analysis?
- What are some practices that you find beneficial while designing, running, or analyzing an experiment with restrictions?
(Note that there is another related discussion started by the same user. I am going to cross-post this call to action and may the best discussion win!)
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: A call to JMP Scientists
Part of the reluctance here might be the language that we use. Perhaps that should have been languages. The scientist talks about plates and wells, pipetting and dilution, incubation time, concentration, and so on. The statistician, on the other hand, (indirectly through the JMP user interface for custom design and directly through replies here) talks about types of factors, easy, hard, or very hard to change factor levels, whole plots and sub-plots, blocking, and so on.
Did you get all that? Probably not. That's normal.
So we need to take the time to explain ourselves to the other people in the discussion. The more advanced the topic and the issues, the more explanation is required to fully communicate the problem and the solution.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: A call to JMP Scientists
Building 96 well experiments.
Take a look at this DOE construction method. There are 4 continuous factors, and 2 additional continuous factors to use as dummy variables for the hard ad very hard to change settings. (dummy variables to construct the whole and subplots.) Factors X3, X4, X5, and X6 are the experimental factos, the design includes interaction and quadratic model terms for each of them. Factors X1 and
X2 are main effects that are set as hard and very hard to change.
(Translated from Stat Speak: If you've got a robot that can pipet a random plate layout, it would be convenient if your DOE table also had plate locations in it too. In this example, I'm going to force JMP to give me plate positions which will be labeled "Subplot" and "Whole Plot." In order to make this work, the first two factors/variables/inputs/X's I add are just going to be dummys. Make the first dummy X hard to change, and make the second X very hard to change. From here add your other variables like drug, inhibitor, buffer, co-factor, nutrient, carbon source or whatever you're interested in studying. Then like normal, add the terms, like interactions and quadratics to the model. Be very careful not to include the dummy X's in the interactions and quadratic terms. All we need are the main effects.
This last step is critical to making this work, don't skip this. There is an option, "Hard to change factors can vary independently of very hard to change factors." This MUST be checked. Take a look at the script and you will notice this: "Make Strip Plot Design", in the arugument. Turning on this option makes that bit occur in the script. (Stats Speak: JMP can make strip plot designs with restrictions on randomization for whole and subplots that are independed of each other.) The next part is important too. Set the number of Whole Plots and Sub Plots, and then the number of runs. One of the plots should be 8 and the other should be 12, and the number of runs is 64. In the past I would have only 8 columns reserved for the experiment, and then use the last three for controls and a standard curves. In this case, Whole Plots and Sub Plots are both 8 and runs = 64. After you make the design table it shouldn't be too hard to pass the design to your robot for pipetting.)
In the design that is generated, JMP provides the 8 whole and 12 subplots in two columns that can be used as the location of each of the 96 wells. In the figure below, X3, X4, X5 and X6 have no restrictions on randomization, and a different pattern is evident for each variable, represented as four the four grids.
After the experiment is complete, it will be important to remember not to include the two dummy variables in the model. By default these will be in the model script.
Design Script
DOE(
Custom Design,
{Add Response( Maximize, "Y", ., ., . ),
Add Factor( Continuous, -1, 1, "whole plot dummy", 1 ),
Add Factor( Continuous, -1, 1, "sub plot dummy", 2 ),
Add Factor( Continuous, -1, 1, "X3", 0 ),
Add Factor( Continuous, -1, 1, "X4", 0 ),
Add Factor( Continuous, -1, 1, "X5", 0 ),
Add Factor( Continuous, -1, 1, "X6", 0 ), Set Random Seed( 42 ),
Number of Starts( 10 ), Add Term( {1, 0} ), Add Term( {1, 1} ),
Add Term( {2, 1} ), Add Term( {3, 1} ), Add Term( {4, 1} ), Add Term( {5, 1} ),
Add Term( {6, 1} ), Add Term( {3, 1}, {4, 1} ), Add Term( {3, 1}, {5, 1} ),
Add Term( {3, 1}, {6, 1} ), Add Term( {4, 1}, {5, 1} ),
Add Term( {4, 1}, {6, 1} ), Add Term( {5, 1}, {6, 1} ), Add Term( {3, 2} ),
Add Term( {4, 2} ), Add Term( {5, 2} ), Add Term( {6, 2} ),
Make Strip Plot Design, Set N Whole Plots( 8 ), Set N Subplots( 12 ),
Set Sample Size( 96 ), Optimality Criterion( 2 ), Make Design}
);
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: A call to JMP Scientists
Thanks for the very helpful input!
Using this, I created a design which would look like this:
Now, varying the cell density on one plate is possible but quite tedious because pipetting cells with a liquid handler needs to be done carefully. In short: I would like to vary the cell density in between plates but not wells. Another factor would be varying incubation time (not shown here), which can also be varied only between plates and not between wells. My approach would be to use the two "dummy" factors as the real deal, namely incubation time and cell density but that does not give the right result. Then, I would also need to take these factors into my model, including interaction and quadratic terms. Where's my mistake? Or how should I approach this?
And, bonus question:
If I want to leave out the edge wells to avoid the edge effect, I would set number of whole plots to 6 and subplots to 10?
Thanks in advance!
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: A call to JMP Scientists
So, this adds a bit more complexity. A practical solution to consider is to repeat the same plate layout for drug, buffer and serum, with different plates of cell concentration. Then to address incubation times, I might prepare multiple plates at each of the cell concentrations so that I could pull plates at appropriate time points.
Of course the the cell conc. will need to be at at least three levels to support a quadratic model term.
Time gets handled differently. I might analyze each time point separately, AND I would fit a curve (Fit Curve) to the response vs. time, outside of the DOE analysis. Then the parameters of the curve (like slope, asymptote, Inflection point/EC50) would become responses in the DOE table. If there isn't already a post on parametizing timeseries data, then that would be a good topic for someone (or me) later.
This can become a fairly large experiment, but that's what pipetting robots are for.
For the bonus question. I expect the change in whole plots and subplots will have little to no effect with so few factors. But like everything, verify first. Make both designs and then use the Design Comparision tool (DOE/Design Diagnostics).
Recommended Articles
- © 2026 JMP Statistical Discovery LLC. All Rights Reserved.
- Terms of Use
- Privacy Statement
- Contact Us