- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Discussions
Solve problems, and share tips and tricks with other JMP users.- JMP User Community
- :
- Discussions
- :
- Re: PCA analysis of time-course data to yield a PCA by Batch ID
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
PCA analysis of time-course data to yield a PCA by Batch ID
Hello,
I have several bioreactor data. Each bioreactor has an ID (1 to 5) and several time-course profile variables (Dissolved oxygen, pH...). I have 20 variables for the time-course data, and 10 timepoints.
When I do a PCA analysis, using the 20 variables as Y columns, I obtain a score plot of the bioreactor data, which is good. Meaning for each bioreactor I get one time-course profile considering the 20 variables, with 10 datapoints per batch. But I would also like to obtain a "Batch ID PCA", in which the data for each bioreactor would be condensed into one datapoint only, and I could identify which bioreactors are similar to each other. SO I would like to get a PCA plot with only 5 datapoints, in which these datapoints would consider the (20*10 data for each bioreactor)
The closer I have been to achieve this was through the K-means cluster platform.
Any help? Preferentially using JMP menus rather than scripting.
I know that one alternative would be to split my matrix. Because the reason I get a time-course data in my PCA analysis is because I have my data table as column 1= ID; Column 2=time; Columns 3-20=Variables.
If I could make only one row to each batch, the PCA analysis would give me what I want. I have tried to split my data, but I have been unsuccessful. If I could convert my data table to column 1= ID; column 2-201 - datapoints, I believe the PCA analysis would give me what I want.
How do I convert my current data table into the 200 new columns? (21 first columns would be time 1, values for the 20 variables for time 1; 21 next columns would be time 2 and the values for the 20 variables for timepoint 2, and so on).
Many thanks
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: PCA analysis of time-course data to yield a PCA by Batch ID
Have you seen what the Functional Data Explorer can do? This documentation includes a biofuels fermentation example that is relevant to your case.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: PCA analysis of time-course data to yield a PCA by Batch ID
Hello,
Thank you for your reply. Unfortunately I do not have JMP Pro. Any other suggestion?
Best,
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: PCA analysis of time-course data to yield a PCA by Batch ID
@DAMP ,
Attached is a subset of the data table Fermentation Process.jmp If you split your Y columns by time (I am assuming you have the same time values), you can create the data table you describe.
Data Table( "Subset of Fermentation Process" ) <<
Split(
Split By( :Time ),
Split( :Ethanol, :Temp, :Molasses Feed, :NH3 Feed, :Air, :Tank Level, :pH ),
Group( :BatchID ),
Remaining Columns( Drop All ),
Sort by Column Property
);However, I am a firm believer of visual analytics and recommend it as the first analysis if your data set is not too large, prior to other modeling.
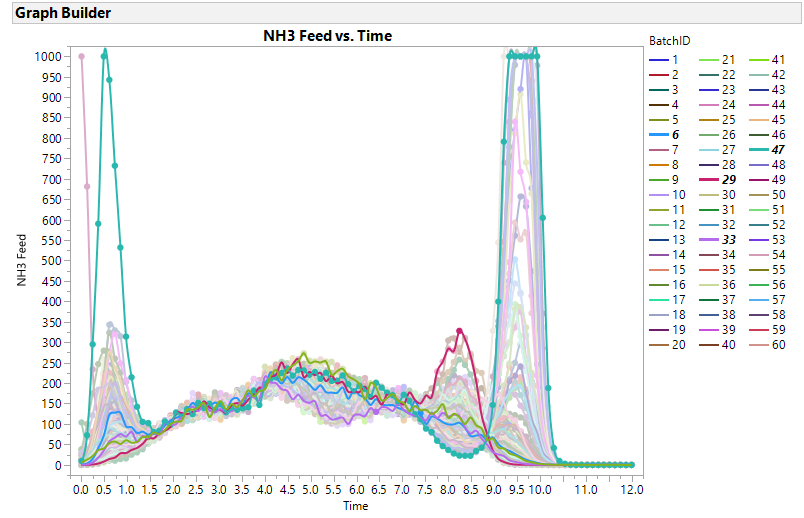
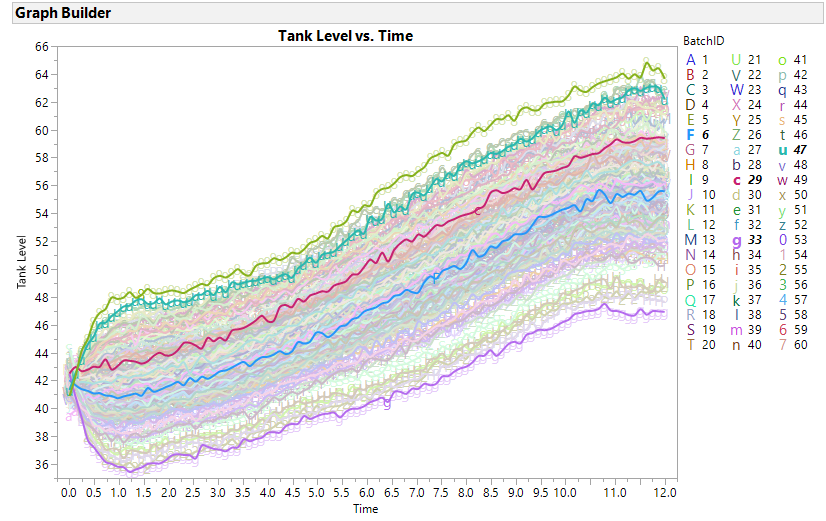
So prior to your PCA, I suggeest you plot each of your 20 variables by time using ID as an Overlay and Color variable in GraphBuilder to get some intuition of the time patterns and the consistency of the reactors. If you had JMP PRO FDE, it creates these plots in addition to methods that can scale and filter and align your data (time warping) which is terrific. However, the visual analysis can potentially lead to more intuitive discriminating variables than eigenvectors. Consider the NH3 Feed vs.Time plot below. BatchID 6, 29, 33 and 47 have different temporal patterns, especially before time 1.5 and after time 7. A metric of start stability and end stability might be better predictors for quality metrics, andoften are more easily explainable to others not versed in PCA.
Just a few suggestions. Good luck!
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: PCA analysis of time-course data to yield a PCA by Batch ID
Hi @gzmorgan0 ,
Thanks so much for your reply!
I appreciate your input regarding the visual analytics. Indeed that is my first step, observing the data and finding odd patterns and profiles.
However for this I would really need to do the PCA as I described, and unfortunately I do not have the same timepoints for each batch. So any other suggestion? Otherwise I guess I will just export the data to excel and do the one batch per row the old, boring way!
Thank you so much for your contribution
Recommended Articles
- © 2026 JMP Statistical Discovery LLC. All Rights Reserved.
- Terms of Use
- Privacy Statement
- Contact Us