- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Discussions
Solve problems, and share tips and tricks with other JMP users.- JMP User Community
- :
- Discussions
- :
- Re: Open multiple Files
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Open multiple Files
I want to open multiple measurement files and concatenate them to a single file.
Unfortunately, due to the strange header of the files, it's not straightforward to use Import Multiple Files ...
So, I set up a for loop and load every file via open() ..., process it one by one, and then concatenate them.
It's astonishing how much slower for( open(...)) is compared to MFI.
Some tricks that I tried:
- load all files, concatenate once --> helps
- option "invisible" (to get rid of table updates) --> helps a lot
but still, no chance to reach the performance of MFI.
Any further tricks?
It should help to use MFI (without stacking), process the data tables separately then and concatenate them.
Is there a command to tell MFI to open the files invisible?
Accepted Solutions
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Open multiple Files
Something like this might work.
// use MFI to stack a mixed bag of tables...
// assumes "text" and "file name" are not columns in the data AND assumes
// no commas in the data values. Not sure how you would work around that.
dir = "$temp/testfiles/"; // make sample files here
Delete Directory( dir );
Create Directory( dir );
Save Text File( dir || "a.csv", "a,b,c\!n1,2,3\!n-1,-2,-3" );
Save Text File( dir || "b.csv", "junk\!nc,b,a,e\!n3,4,5,98\!n-1,-2,-3,-4" );
Save Text File( dir || "c.csv", "junk\!njunk\!nb,d,a,c\!n6,99,7,8\!n-1,-2,-3,-4" );
dtList = Multiple File Import(
<<Set Folder( dir ),
<<Set Add File Name Column( 1 ),
<<Set Import Mode( "Row Per Line" ),
<<Set Stack Mode( "Stack Similar" )
) << Import Data;
dt = dtList[1];
// clean up "junk". It might not be this simple for your data...
dt << selectwhere( Contains( text, "junk" ) );
dt << deleterows;
// hunt for unique column names
headerrows = dt << getrowswhere( Row() == 1 | FileName[Row()] != FileName[Row() - 1] );
uniquecols = Associative Array( Words( Concat Items( Transform Each( {header}, dt[headerrows, {text}], header[1] ), "," ), "," ) ) << getkeys;
// add the unique column names, character data for now...
For Each( {cname}, uniquecols, dt << New Column( cname, character ) );
// transfer data from text to unique cols.
headerrows |/= N Rows( dt ) + 1; // append sentinel
For( iheader = 1, iheader < N Rows( headerrows ), iheader += 1,
cols = Words( dt:text[headerrows[iheader]], "," );
startrow = headerrows[iheader] + 1;
stoprow = headerrows[iheader + 1] - 1;
For( irow = startrow, irow <= stoprow, irow += 1,
dt[irow, cols] = Words( dt:text[irow], "," )
);
);
// remove the headers and left over bits
dt << selectrows( headerrows[1 :: N Rows( headerrows ) - 1] );
dt << deleterows;
dt << delete columns( {text, file name} );
dt << delete scripts( "files" );
dt << delete scripts( "source" );
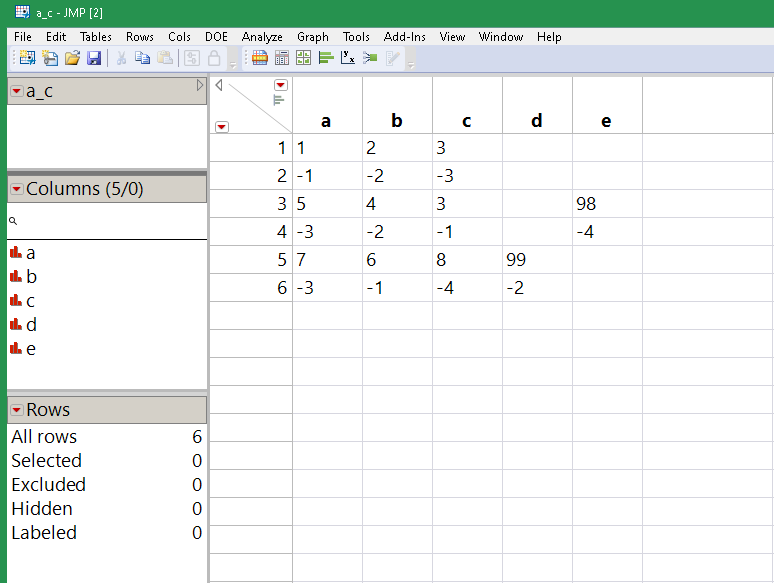
Now you'll have to decide which columns are numeric...
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Open multiple Files
How about this:
- use the ultra-fast MFI-Import (with csv-option)
- separate the input files via Subset(private)
- postprocess the data to get meaningful column names [here just a moveup]
- concatenate the files
// use MFI to stack a mixed bag of tables...
// assumes "text" and "file name" are not columns in the data AND assumes
// no commas in the data values. Not sure how you would work around that.
// then splits the data via Subset(private)
// ... some postprocessing ...
// and then concatenate the files
dir = "$temp/testfiles/"; // make sample files here
open(dir);
Delete Directory( dir );
Create Directory( dir );
dt0 = Open( "$SAMPLE_DATA/Big Class.jmp" );
Foreach ({i,idx},1::300,
dt0 << Save( dir || char(i)||".csv" ));
t1=HPtime();
dtList = (Multiple File Import(
<<Set Folder( dir ),
<<Set Add File Name Column( 1 ),
<<Set Import Mode( "CSVData" ),
<<Set Stack Mode( "Stack Similar" ),
<<Set CSV Has Headers( 0 ),
<<Set CSV Allow Numeric( 0 ),
<<Set CSV First Data Line( 1 ),
<<Set CSV EOF Comma( 1 )
) << Import Data)[1];
t2 = HPtime();
myDTs =dtList << Subset(By( :File Name ), Private);
Close(dtList,noSave);
myDTs << Move up;
t3 = HPtime();
myDTs[1] << Concatenate( myDTs[2::N Items(myDTs)], Append to first table ); // geht auch, wenn n=1
finalVersion = myDTs[1] << Subset( Link To Original Data Table( 0 ), Output Table( Word(-2,dir,"\/") ));
For Each( {dt}, myDTs, Close( dt, noSave ) );
t4 = HPtime();
show((t2-t1)/1000000,(t3-t2)/1000000,(t4-t3)/1000000)
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Open multiple Files
Good catch, the setup for the data table matrix columns is pretty steep. This way uses one setup for all 40 rows instead of one for every row. (see the temp list.) I see ~4X better for the big class test case:
// use MFI to stack a mixed bag of tables...
// assumes "text" and "file name" are not columns in the data AND assumes
// no commas in the data values. Not sure how you would work around that.
dir = "$temp/testfiles/"; // make sample files here
Open( dir );
Delete Directory( dir );
Create Directory( dir );
dt0 = Open( "$SAMPLE_DATA/Big Class.jmp" );
For Each( {i, idx}, 1 :: 300,
dt0:height[1] = 1000 + i;
dt0 << Save( dir || Char( 1000+i ) || ".csv" );
);
t1 = HP Time();
dtList = Multiple File Import(
<<Set Folder( dir ),
<<Set Add File Name Column( 1 ),
<<Set Import Mode( "Row Per Line" ),
<<Set Stack Mode( "Stack Similar" )
) << Import Data;
dt = dtList[1];
t2 = HP Time();
//dtList = dtList << subset(private);
// clean up "junk". It might not be this simple for your data...
dt << selectwhere( Contains( text, "junk" ) );
dt << deleterows;
// hunt for unique column names
headerrows = dt << getrowswhere( Row() == 1 | FileName[Row()] != FileName[Row() - 1] );
uniquecols = Associative Array( Words( Concat Items( Transform Each( {header}, dt[headerrows, {text}], header[1] ), "," ), "," ) ) << getkeys;
// add the unique column names, character data for now...
For Each( {cname}, uniquecols, dt << New Column( cname, character ) );
// transfer data from text to unique cols.
headerrows |/= N Rows( dt ) + 1; // append sentinel
For( iheader = 1, iheader < N Rows( headerrows ), iheader += 1,
cols = Words( dt:text[headerrows[iheader]], "," );
startrow = headerrows[iheader] + 1;
stoprow = headerrows[iheader + 1] - 1;
temp = {}; // create a bigger block of rows...
For( irow = startrow, irow <= stoprow, irow += 1,
temp[N Items( temp ) + 1] = Words( dt:text[irow], "," )
);
dt[startrow :: stoprow, cols] = temp; // ...and do the column lookup once
);
// remove the headers and left over bits
dt << selectrows( headerrows[1 :: N Rows( headerrows ) - 1] );
dt << deleterows;
dt << delete columns( {text, file name} );
dt << delete scripts( "files" );
dt << delete scripts( "source" );
//dtList << subset(visible);
t3 = HP Time();
Show( (t2 - t1) / 1000000, (t3 - t2) / 1000000 );(t2 - t1) / 1000000 = 2.105032;
(t3 - t2) / 1000000 = 2.65675;
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Open multiple Files
- Seems like some good wish list items: a more aggressive concatenate option, an invisible option.
- try minimizing the tables
- try <<BeginDataUpdate
- MFI also has a faster CSV reader than open() uses. But using open can still be fast if you can use the source script that specifies enough information to prevent an extra scan of the data. Or try using MFI to import a single file.
- MFI uses multiple threads. If you are looking at minutes of time (not seconds), Can JMP run JMP? might be what you need to run more than one open() at a time. It isn't clear that the time you spend making it work would ever pay back, and a nightmare to maintain for someone else.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Open multiple Files
While writing the post for the wish list, I found related wishes - and added links there:
Invisible: there is already a wish to open the files with private option (from 2022):
Private option for Import Multiple Files
aggressive concatenation (from 2020):
Multiple File Import - Force Combine
So, with some luck they are already implemented in the next version of Jmp :)
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Open multiple Files
I have written an addin that does this with a few other options and works soley on the power of MFI. I haven't shared it on the community as I need to scrub some company specific items from it. This may spure that to share what i've learned over the years :)
Anyways, you can use MFI and what I do is basically create a list of the file names (via pick file or files in directory) so I have a singular path and a list of file names for that path, and then pass them into the MFI command.
Then I check the output and if it's more than 1 file I'll force concatenate. Here's a snipit
filter = ConcatItems(fileList,";");
filesImported1 = Multiple File Import(
<<Set Folder( path ),
<<Set Name Filter( filter ),
<<Set Name Enable( 1 ),
<<Set Add File Name Column(1),
<<Set Import Mode("CSV Data"),
<<Set Stack Mode("Stack Similar")
)<<Import Data();
filesImported1 << show window(0);filter is basically the list of file names, but separated by ";"
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Open multiple Files
Hi @nathan-clark .
thank you for the sniplet.
Concerning the application case discussed here, some preprocess on file level is necessary before the files can be merged.
Therefore, I would prefer to load the files with the option
<<Set Stack Mode( "Table Per File" )
On my system (Jmp 17.1), Jmp starts to display the 100+ tables before it reacts on the
<< show window(0);
This was the reason for:
@hogi wrote:Is there a command to tell MFI to open the files invisible?
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Open multiple Files
Ok that makes more sense. I see what you are getting at...
The only thing that will make it a bit faster is if you open the files privately instead of invisible. It's quite a speedy upgrade... not sure if it will match the speed of MFI though. I've found that even when private, JMP can do most operations on the files and it drastically reduces RAM needs... you just need to comment out private when testing or you end up looking for test tables you can't see :)
Hopefully JMP can improve MFI even further.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Open multiple Files
@nathan-clark , thanks a lot, the private makes it roughly factor 2 faster :)
@nathan-clark wrote:Hopefully JMP can improve MFI even further.
Yes, an invisible (or even private!) option in MFI
https://community.jmp.com/t5/JMP-Wish-List/Private-option-for-Import-Multiple-Files/idc-p/632087#M42...
will be a GameChanger !
Enough options for me:
- use the private mode in Open and take a short nap
- check if I manage to adapt @Craige_Hales 's solution below to our file type.
- wait for the invisible option in Jmp 18 ;)
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Open multiple Files
Something like this might work.
// use MFI to stack a mixed bag of tables...
// assumes "text" and "file name" are not columns in the data AND assumes
// no commas in the data values. Not sure how you would work around that.
dir = "$temp/testfiles/"; // make sample files here
Delete Directory( dir );
Create Directory( dir );
Save Text File( dir || "a.csv", "a,b,c\!n1,2,3\!n-1,-2,-3" );
Save Text File( dir || "b.csv", "junk\!nc,b,a,e\!n3,4,5,98\!n-1,-2,-3,-4" );
Save Text File( dir || "c.csv", "junk\!njunk\!nb,d,a,c\!n6,99,7,8\!n-1,-2,-3,-4" );
dtList = Multiple File Import(
<<Set Folder( dir ),
<<Set Add File Name Column( 1 ),
<<Set Import Mode( "Row Per Line" ),
<<Set Stack Mode( "Stack Similar" )
) << Import Data;
dt = dtList[1];
// clean up "junk". It might not be this simple for your data...
dt << selectwhere( Contains( text, "junk" ) );
dt << deleterows;
// hunt for unique column names
headerrows = dt << getrowswhere( Row() == 1 | FileName[Row()] != FileName[Row() - 1] );
uniquecols = Associative Array( Words( Concat Items( Transform Each( {header}, dt[headerrows, {text}], header[1] ), "," ), "," ) ) << getkeys;
// add the unique column names, character data for now...
For Each( {cname}, uniquecols, dt << New Column( cname, character ) );
// transfer data from text to unique cols.
headerrows |/= N Rows( dt ) + 1; // append sentinel
For( iheader = 1, iheader < N Rows( headerrows ), iheader += 1,
cols = Words( dt:text[headerrows[iheader]], "," );
startrow = headerrows[iheader] + 1;
stoprow = headerrows[iheader + 1] - 1;
For( irow = startrow, irow <= stoprow, irow += 1,
dt[irow, cols] = Words( dt:text[irow], "," )
);
);
// remove the headers and left over bits
dt << selectrows( headerrows[1 :: N Rows( headerrows ) - 1] );
dt << deleterows;
dt << delete columns( {text, file name} );
dt << delete scripts( "files" );
dt << delete scripts( "source" );

Now you'll have to decide which columns are numeric...
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Open multiple Files
Thanks @Craige_Hales - many really good ideas to copy!
Unfortunately, the
// transfer data from text to unique cols.
...is quite slow ...
here an example - on my system it takes one second to load the data and 10 seconds to convert the data ...
// use MFI to stack a mixed bag of tables...
// assumes "text" and "file name" are not columns in the data AND assumes
// no commas in the data values. Not sure how you would work around that.
dir = "$temp/testfiles/"; // make sample files here
open(dir);
Delete Directory( dir );
Create Directory( dir );
dt0 = Open( "$SAMPLE_DATA/Big Class.jmp" );
Foreach ({i,idx},1::300,
dt0 << Save( dir || char(i)||".csv" ));
t1=HPtime();
dtList = Multiple File Import(
<<Set Folder( dir ),
<<Set Add File Name Column( 1 ),
<<Set Import Mode( "Row Per Line" ),
<<Set Stack Mode( "Stack Similar" )
) << Import Data;
dt = dtList[1];
t2=HPtime();
//dtList = dtList << subset(private);
// clean up "junk". It might not be this simple for your data...
dt << selectwhere( Contains( text, "junk" ) );
dt << deleterows;
// hunt for unique column names
headerrows = dt << getrowswhere( Row() == 1 | FileName[Row()] != FileName[Row() - 1] );
uniquecols = Associative Array( Words( Concat Items( Transform Each( {header}, dt[headerrows, {text}], header[1] ), "," ), "," ) ) << getkeys;
// add the unique column names, character data for now...
For Each( {cname}, uniquecols, dt << New Column( cname, character ) );
// transfer data from text to unique cols.
headerrows |/= N Rows( dt ) + 1; // append sentinel
For( iheader = 1, iheader < N Rows( headerrows ), iheader += 1,
cols = Words( dt:text[headerrows[iheader]], "," );
startrow = headerrows[iheader] + 1;
stoprow = headerrows[iheader + 1] - 1;
For( irow = startrow, irow <= stoprow, irow += 1,
dt[irow, cols] = Words( dt:text[irow], "," )
);
);
// remove the headers and left over bits
dt << selectrows( headerrows[1 :: N Rows( headerrows ) - 1] );
dt << deleterows;
dt << delete columns( {text, file name} );
dt << delete scripts( "files" );
dt << delete scripts( "source" );
//dtList << subset(visible);
t3 = HPtime();
show((t2-t1)/1000000,(t3-t2)/1000000)
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Open multiple Files
How about this:
- use the ultra-fast MFI-Import (with csv-option)
- separate the input files via Subset(private)
- postprocess the data to get meaningful column names [here just a moveup]
- concatenate the files
// use MFI to stack a mixed bag of tables...
// assumes "text" and "file name" are not columns in the data AND assumes
// no commas in the data values. Not sure how you would work around that.
// then splits the data via Subset(private)
// ... some postprocessing ...
// and then concatenate the files
dir = "$temp/testfiles/"; // make sample files here
open(dir);
Delete Directory( dir );
Create Directory( dir );
dt0 = Open( "$SAMPLE_DATA/Big Class.jmp" );
Foreach ({i,idx},1::300,
dt0 << Save( dir || char(i)||".csv" ));
t1=HPtime();
dtList = (Multiple File Import(
<<Set Folder( dir ),
<<Set Add File Name Column( 1 ),
<<Set Import Mode( "CSVData" ),
<<Set Stack Mode( "Stack Similar" ),
<<Set CSV Has Headers( 0 ),
<<Set CSV Allow Numeric( 0 ),
<<Set CSV First Data Line( 1 ),
<<Set CSV EOF Comma( 1 )
) << Import Data)[1];
t2 = HPtime();
myDTs =dtList << Subset(By( :File Name ), Private);
Close(dtList,noSave);
myDTs << Move up;
t3 = HPtime();
myDTs[1] << Concatenate( myDTs[2::N Items(myDTs)], Append to first table ); // geht auch, wenn n=1
finalVersion = myDTs[1] << Subset( Link To Original Data Table( 0 ), Output Table( Word(-2,dir,"\/") ));
For Each( {dt}, myDTs, Close( dt, noSave ) );
t4 = HPtime();
show((t2-t1)/1000000,(t3-t2)/1000000,(t4-t3)/1000000)
Recommended Articles
- © 2026 JMP Statistical Discovery LLC. All Rights Reserved.
- Terms of Use
- Privacy Statement
- Contact Us


