- JMP User Community
- :
- Discussions
- :
- Module analysis and correlation networks
- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Module analysis and correlation networks
This might be more of a "wish list" post, but I am interested in the capacity of carrying out correlation network analysis in JMP with OMICS datasets (in my case, protein concentration data). There is a popular approach called WGCNA (weighted gene co-expression network analysis), described as follows (from https://horvath.genetics.ucla.edu/html/CoexpressionNetwork/Rpackages/WGCNA/):
"Correlation networks are increasingly being used in bioinformatics applications. For example, weighted gene co-expression network analysis is a systems biology method for describing the correlation patterns among genes across microarray samples. Weighted correlation network analysis (WGCNA) can be used for finding clusters (modules) of highly correlated genes, for summarizing such clusters using the module eigengene or an intramodular hub gene, for relating modules to one another and to external sample traits (using eigengene network methodology), and for calculating module membership measures. Correlation networks facilitate network based gene screening methods that can be used to identify candidate biomarkers or therapeutic targets. These methods have been successfully applied in various biological contexts, e.g. cancer, mouse genetics, yeast genetics, and analysis of brain imaging data. While parts of the correlation network methodology have been described in separate publications, there is a need to provide a user-friendly, comprehensive, and consistent software implementation and an accompanying tutorial."
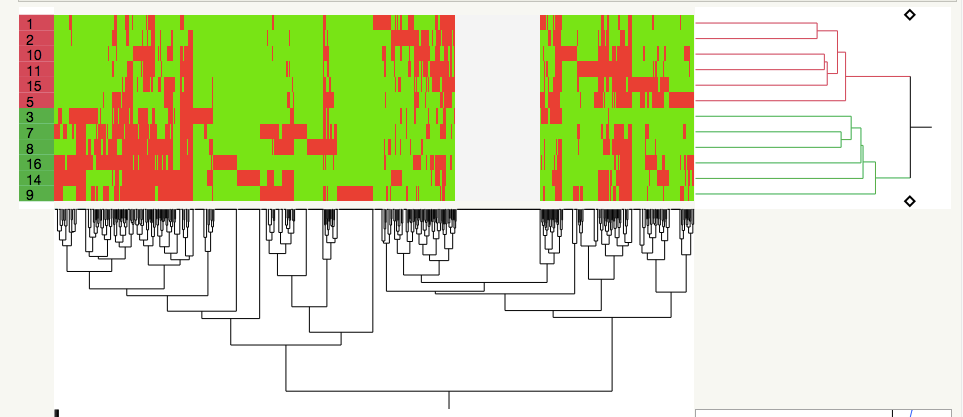
It seems to me that this is just a fancy way of clustering by covariance, which I DO know how to do with JMP Pro 14 (I've looked at clustering across my sample set=biological replicates alongside clustering of the various proteins whose concentrations I measured in each sample). Basically, I like what the authors have done in the first figure I've attached: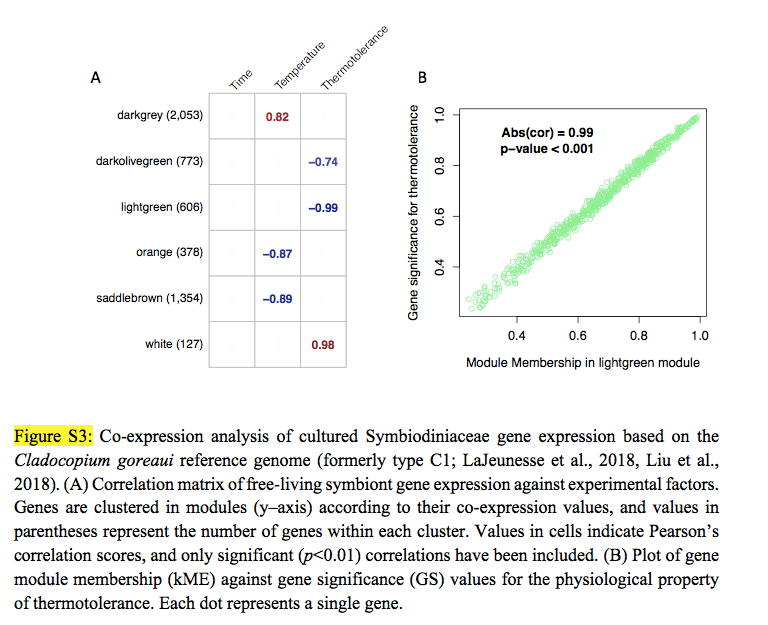
the samples (left side) are in two clusters based on their proteome profiles, whereas on the x axis, you can see 10-12 general clusters of proteins (I could transpose the dataset and have each protein assigned a cluster).
Basically, I want to look at module (cluster?) correlations across temperature, time, and genotype in this attached dataset to where I can identify clusters of proteins that correlate with these experimental factors of interest. It actually seems like partial least squares could be used for this, too.....Any ideas out there?
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Module analysis and correlation networks
Nice analyses @abmayfield--you have that JMP table tricked out! With only 12 observations and binary presence/absence for the 769 proteins, it’s important not to overfit or overinterpret, but here are several more ideas:
- The following steps should produce a WGCNA-style analysis:
1. Transpose, delete the four poor-quality samples
2. Hierarchical Clustering, two-way to get heatmap like you have already done
3. Choose the number of clusters interactively (coloring can help) and Save Cluster as a new column
4. Summarize to get the mean of all variables by Cluster, which is an average protein profile for each cluster, aka eigenprotein. When running Summary, specify “column” as “statistics column name format” to facilitate the next step.
5. Transpose back and merge with experimental factors
6. Create numerical versions of the experimental factors
7. Multivariate
- More principled and powerful is to fit ANOVA models with all three factors at once. Need to be careful with limited degrees of freedom. You can do this with the eigenproteins and Fit Model. You can also do it with the original proteins but the large number of reports can be unwieldy. The main trick in this case is to right click on any report table > Make Combined Data Table.
- JMP Genomics has a more comprehensive workflow and interactive dashboard output from its Row-by-Row Modeling menu. You can even do mixed models (e.g. make genotype a random effect). JMP Genomics has numerous other helpful routines, as it is designed for high-throughput data sets like this one.
- Use significant proteins of interest and/or the preceding eigenproteins in Structural Equation Modeling or the Partial Correlation Diagram add-in to infer potential causal relationships
- If you have pathway annotations for the proteins, compute pathway-based scores and then analyze them versus the experimental factors.
- Try the add-in from MJ Guan for low-dimensional projections based on t-SNE and UMAP.
- For PLS I think you would first need to create binary indicator variables for all experimental factors with Cols > Utilities > Make Indicator Columns
- Run Analyze > Screening > Response Screening to fit all Y by X combos and select proteins based on FDR-adjusted p-values. I tried this and nothing is statistically significant, but there is a dozen or so proteins with small raw p-values.
- If you can obtain continuous measures of protein expression instead of presence/absence the preceding analyses should be more informative.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Module analysis and correlation networks
Russ,
Wow, thank you for thinking this through so carefully. I now have similar data, but fully quantitative (see attached). One issue might be, though, is that these proteomic datasets are much smaller than their mRNA counterparts. In this example, there are only 40 proteins (though it can be in the hundreds depending on the sample). But, if nothing else, I can at least go through the pipeline you have nicely drawn out. It does get to what was my real question, which was: is WGCNA really just regressing mean values from clusters of molecules against physiological parameters, to which, according to your response, that is indeed basically what it is. I am going to go through this today and report back to here with questions/comments/etc. Thanks again for your help and stay tuned for more.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Module analysis and correlation networks
If JMP Genomics can really do these such analyses, I may need to explore it in more detail.....
- © 2024 JMP Statistical Discovery LLC. All Rights Reserved.
- Terms of Use
- Privacy Statement
- About JMP
- JMP Software
- JMP User Community
- Contact