- We’re retiring the File Exchange at the end of this year. The JMP Marketplace is now your destination for add-ins and extensions.
- JMP 19 is here! Learn more about the new features.
- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Discussions
Solve problems, and share tips and tricks with other JMP users.- JMP User Community
- :
- Discussions
- :
- Re: How to see Fit Definitive Screening Design active effect decisions by stage?
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
How to see Fit Definitive Screening Design active effect decisions by stage?
Using JMP 17.0, is there any way to see how it picked the effects it picked as active in the Fit Definitive Screening Report?
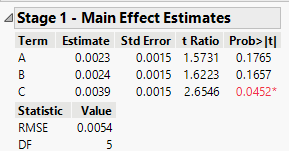
For example, the Help file says in Stage 1 with fake factor or center point replicates, an estimator of error variance independent of the model is constructed. Main effects are tested against this estimate and those with p-values less than a threshold p-value are considered active. The threshold value, unless set manually, varies by degrees of freedom. Where is this process shown?
Is that estimate the reported RMSE and the degrees of freedom below it? If so, then where are the p-values for each main effect so I can see what JMP estimates them as and how they are less than the threshold p-value for each main effect it picked? I don't see them. Instead I get a table of t Ratios and Prob > |t| and that table shows active effects for prob > |t| much larger than the p-value of 0.05 -- like a probability of 0.18, that suggests to me this Prob > |t| is not the p-value it compares to the threshold to determine which effects are active.
I'd even like to see what it estimated for all that it didn't pick as active -- didn't meet the criteria. By how much?
And the same question for Stage 2.
Sorry, my knowledge and skill involves reading a standard ANOVA table from factorial or central composite designs. I got JMP because of the special method it uses to analyze DSDs, but I am having trouble learning how to read and understand the results it gives. Stepwise analysis of my DSD gives a very different answer, picks many more effects with "Prob>F" less than 0.05., using AICc or BIC. I'm trying to reconcile the two and most importantly, understand the results for the Fit Definitive Screening Design report (prior to run or make model) that is so new to me.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: How to see Fit Definitive Screening Design active effect decisions by stage?
Not certain I understand your question, but in the help (as you say) it says further:
"Using YME, main effects are tested against this estimate. Main effects with p-values less than a threshold p-value are considered active. The threshold values are the following:
–For one error degree of freedom, the threshold value is 0.20.
–For two error degrees of freedom, the threshold value is 0.10.
–For more than two error degrees of freedom, the threshold value is 0.05."
In your case, DF=5 so the threshold is 0.05. That is why factor C is indicated as "active" (its p-value, Prob >|t|, is less than 0.05).
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: How to see Fit Definitive Screening Design active effect decisions by stage?
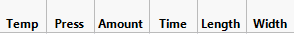
There are 6 factors in the DSD. The table shows three of them, highlighting one of the three, and not showing the other three at all. Why those three? And where are the other three? And why are two of them listed with Prob >|t| well above 0.05? And the three missing are not shown?
I thought it was only showing those where the p value for the effect was less than the threshold p-value. If Prob >|t| is the p value, then it is showing two active effects that do not meet that threshold. And it doesn't show the other three at all. This suggests to me Prob >|t| is NOT the p value for the effect.
In that case, where is the p value for the effect that it is using to determine if that effect is less than the threshold p-value? That's my question. How do I see those p values for each effect so I can inspect that it only picked those less than the threshold p-value. And I'd like to see the other p values so I can see how far they were from being picked as active.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: How to see Fit Definitive Screening Design active effect decisions by stage?
Hmmm...if you have 6 main effects (A, B, C, D, E, F, rather than three main effects and three 2nd order terms: A, B, C, AB, AC, BC or A, B, C, A^2, B^2, C^2) then I'm not sure what is going on.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: How to see Fit Definitive Screening Design active effect decisions by stage?
Yes, sorry, I should have said in my original question it is a 6 factor 17 run DSD depicted in the picture, but my specific DSD is not pertinent to my question.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: How to see Fit Definitive Screening Design active effect decisions by stage?
What I suspect is the answer:
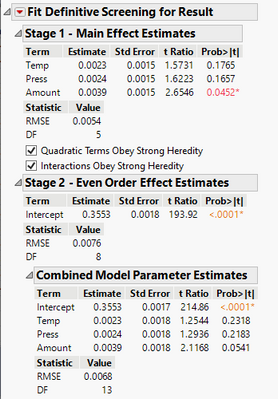
Because I had fake factors (4 extra runs) in my one example, JMP first computed an estimate of the error variance independent of the model. It then used the YME, which I think is the first column in the Stage 1 output table, and "tested them against this estimate". I don't know how many degrees of freedom it had for this and I'm puzzled by the fact that if I specify the p-value threshold to 0.2, two of the odd order effects (main effects) drop from the output table. So I suspect it was using 0.05 and the error variance had more than 2 degrees of freedom. So it then concluded three of the six odd order effects (main effects) were below this 0.05 threshold, the other three not, so it holds on to the three that were below 0.05.
It then took the variability from the other three main effects and pooled them into the error variance, changing the standard errors, the t-ratios and the p value in the prob > |t| column, which it says in Help that it is the p-value. So the p-values for two of the six factors are no longer below the threshold because of this pooling of the additional degrees of freedom into the error variance changed the error variance and every resultant calculation. And the RMSE shown is this final error variance estimate? The original one before the pooling is not shown and it appears there is no option anywhere to show it. I'd really like to see it -- or rather, the entire table for all the main effects before the decision what to keep and what not to keep and before the pooling of those not kept into error.
If I am correct, it is interesting the table then shows them as they are no longer with a p-value below 0.05. It then keeps them. for the Combined Model Parameter Estimates table after Stage 2.
So I"m not sure how to interpret them. The clean notion that those main effects with a p-value below 0.05 are "active effects" and those not are not active turns into an even more murky grey. In my example those with Prob > |t| of 0.1765 and 0.1657 are active effects before the pooling of additional degrees of freedom into error, making them what? Lazy effects? Ineffective effects? I like that one. :) I suspect better names would be obvious effects and non-obvious possible effects.
Perhaps JMP keeps them because they originally had p-values below 0.05, so it might be wise to keep them in subsequent non-screening experimentation.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: How to see Fit Definitive Screening Design active effect decisions by stage?
Hi @CanonicalHazard ,
Sorry, I'm struggling to follow this thread and understand all the specific questions you are trying to answer. Attaching illustrative example data really helps us to give specific answers.
But I think that your "What I suspect is the answer" sounds about right. The method is not simple but it seems fairly well documented to me. If there is something in the documentation that is not clear or that you think is missing or incorrect then you might want to contact JMP technical support (support@jmp.com). Again, illustrative example data will be useful.
How to interpret the result? I think that you are on the right lines with this as well.
DSDs are screening designs, so the objective is to understand the factors that have an important effect on the response(s) to carry forward for further study in the next stages of your sequence of experiments.
Unfortunately there is no such thing as a "clean notion" of what defines an "active" effect or factor. Personally I am not very comfortable with any distinction between active and inactive, it's just wrong to me to make that distinction. And I am certainly not comfortable that p < 0.05 is a clean distinction.
Fit DSD is a useful and quick way to find out which factors and effects are most important.
But if you want an analysis for interpretation, particularly if you are publishing, you might want to stick to the traditional model selection methods as these are better known.
You will get different results between model selection methods and that is not a bad thing. If there is reasonable consensus, then it provides useful confirmation that you can be confident of the result.
If I see big differences then I seek to understand more about the differences and what the ambiguity is. I look at more than just p-values. In fact, I don't look much at p-values. I would spend more time looking at the prediction profiler as I am much more interested in the directions and sizes of effects. When there are important differences I can use that to guide the collection of more data to get closer to resolving the ambiguities.
If you have suggestions for improving the Fit DSD report you can add them to the JMP Wish List.
I hope this helps,
Phil
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: How to see Fit Definitive Screening Design active effect decisions by stage?
Here is the example, with made up factor names. Six factors. The process is a Finite Element Analysis simulation (zero replication error).
It is a 17 run DSD.
I analyze it with "Fit Definitive Screening Design"
I get this
You can see three of the six factors are presented in the Stage 1 table.
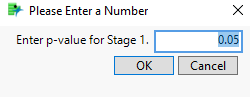
The Stage 1 p-value setting is
Question: Why are Temp and Press listed in the Stage 1 table along with Amount? The Prob > |t| is NOT below 0.05 for Temp and Press.
Question 2: Why were Time, Length, Width not listed in the Stage 1 table?
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: How to see Fit Definitive Screening Design active effect decisions by stage?
You might be interested in the original paper describing the method implemented in the Fit Definitive Screening procedure in JMP.
Bradley Jones & Christopher J. Nachtsheim (2016): Effective Design-Based Model Selection for Definitive Screening Designs, Technometrics, DOI: 10.1080/00401706.2016.1234979
Second, other model selection methods might be more satisfying because they act less like a 'black box.'
Third, your last example has no random error in it. Most of the design methods in JMP try to achieve an optimal design that minimizes a standard error associated with a model. The finite precision numerical methods used to estimate model parameters and standard errors often fail when the sum of squares error is zero. (It's not math, it's computing.) You should not have any such error. A computer simulation like yours is usually approached with a space-filling design and then modeled with a Gaussian Process for rapid interpolation. The idea of screening or determining statistical significance is confusing when you can look at the simulation code or have no stochastic element.
Recommended Articles
- © 2025 JMP Statistical Discovery LLC. All Rights Reserved.
- Terms of Use
- Privacy Statement
- Contact Us