- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Discussions
Solve problems, and share tips and tricks with other JMP users.- JMP User Community
- :
- Discussions
- :
- Re: How to focus LSMeans Comparisons in the Fit Model report for an interaction ...
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
How to focus LSMeans Comparisons in the Fit Model report for an interaction term with large number of levels?
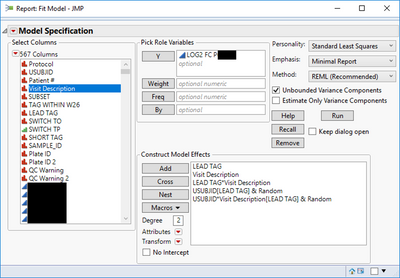
I have analyzed a biomarker (LOG2 FC P) data set using a Repeat Measure linear fit model as shown below
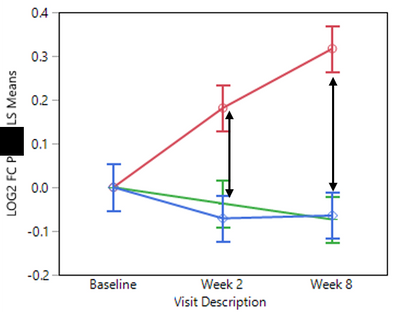
Now, I would like to compare the treatments (LEAD_TAG) at each time point (Visit_Description) without generating all possible comparison pairs produced by the LSMEans Differences Tukey HSD method (see graph below):
- is it possible to do so?
- Does it matter? (i.e. is there a "cost" for comparing all pairs [36 comparisons] instead of the desired ones [6 comparisons])
Accepted Solutions
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: How to focus LSMeans Comparisons in the Fit Model report for an interaction term with large number of levels?
Hi @Thierry_S ,
There is a cost in using the Tukey HSD pairwise method since the correction applied to keep your overall alpha at the stated level takes into account the number of comparisons. If you need to look at only 3, you're essentially paying with statistical power by correcting for 36.
I would use one of the LSMeans Contrast options, right next toTukey HSD in the Red Triangle for your interaction between treatment and timepoint. You could either specify the linear contrasts yourself by selecting LSMeans Contrast three times, or by using "Test Slices," which will automatically test each factor at a slice along the other factor. In your case, that means you will get 6 tests: three tests of time, one for each treatment, and three tests of treatment, one for each time. It's the last three you want, and they'll be labeled accordingly (baseline, week 2, week 8), and they test whether you have evidence there is an effect of treatment at the given slice of time.
Note that these tests will not correct for multiple comparisons -- the p-values reported are unadjusted. With three planned comparisons, a Bonferroni correction will likely be most useful.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: How to focus LSMeans Comparisons in the Fit Model report for an interaction term with large number of levels?
Hi @Thierry_S ,
There is a cost in using the Tukey HSD pairwise method since the correction applied to keep your overall alpha at the stated level takes into account the number of comparisons. If you need to look at only 3, you're essentially paying with statistical power by correcting for 36.
I would use one of the LSMeans Contrast options, right next toTukey HSD in the Red Triangle for your interaction between treatment and timepoint. You could either specify the linear contrasts yourself by selecting LSMeans Contrast three times, or by using "Test Slices," which will automatically test each factor at a slice along the other factor. In your case, that means you will get 6 tests: three tests of time, one for each treatment, and three tests of treatment, one for each time. It's the last three you want, and they'll be labeled accordingly (baseline, week 2, week 8), and they test whether you have evidence there is an effect of treatment at the given slice of time.
Note that these tests will not correct for multiple comparisons -- the p-values reported are unadjusted. With three planned comparisons, a Bonferroni correction will likely be most useful.
Recommended Articles
- © 2026 JMP Statistical Discovery LLC. All Rights Reserved.
- Terms of Use
- Privacy Statement
- Contact Us