For context, I am running analyses on a proteomics experiment. I have two datasets: Intraday (comparing AM to PM), and Longitudinal (comparing Day 0 to Day 30 to Day 90) for protein concentration. For Intraday, I have 600 proteins I am looking at. For Longitudinal, I have 68. I want to find out the magnitude of change between the time points for each protein, so I am trying to find z-scores. I am new to statistical analyses as I am a student who is new to this kind of research, so I am not completely sure what I am doing is right.
To find Z-scores for the Intraday dataset, I input the "Intraday" data, in the attached excel below, into JMP. The JS value is Johnson transformed values for normality. I then followed these steps:
- Take Johnson Transformed Data --> Fit Y by X --> Y = JS value, X = time, By = Protein Name --> Get graphs --> Red Triangle --> Nonparametric --> Exact Test --> Wilcoxin Exact Test
- My data looked like this:
-
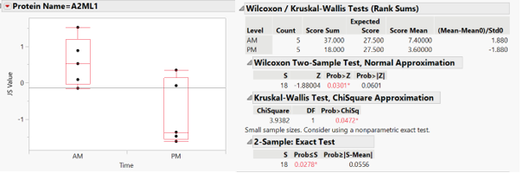
- From there, I did: See Wilcoxin 2-sample test, normal approx. --> right click --> Make combined table --> See z-scores --> Paste into Excel
- Then my data looked like this:
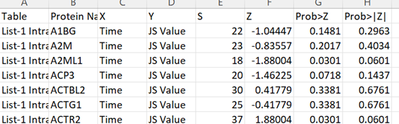
- Here are some of the z-scores for the Intraday data under the "Z" tab.
My issues is that I cannot perform the same method to get z-scores for my longitudinal data, likely because I am now comparing 3 time points instead of 2, so I cannot do a simple Fit Y by X (that I know of). I would appreciate any and all feedback as to how to operate JMP into getting me Z-scores for each protein like you see above for Intraday, but for the Longitudinal dataset.
Thank you so much