- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Discussions
Solve problems, and share tips and tricks with other JMP users.- JMP User Community
- :
- Discussions
- :
- Re: Creating a Venn diagram based on protein frequency
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Creating a Venn diagram based on protein frequency
Hello all,
I have run into a quandary, and I feel confident there is a simple solution. I have sequenced several hundred proteins for three samples (actually, it's more, but let's start easy): A4-1, A4-5, and A4-8 I want to create a Venn diagram to depict the proteins that were uniquely expressed by certain samples vs. those that were shared. I have now essentially stacked all of the protein names into one column. The problem is, if I try to transpose to where the protein names are in the columns, it then merges those with the same name, which is NOT what I want. If I give each column a unique label, though, I need to go through and manually merge them, which could take a long time. I would like an output as in the attached image, with 0s and 1s represent absence and presence, respectively, of the protein listed in the column. 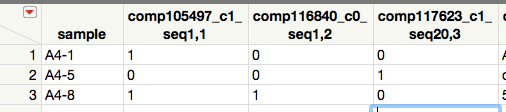
Accepted Solutions
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Creating a Venn diagram based on protein frequency
I do not understand your data, especially how time and treatment should be factored into the analysis. I think Tables> Summary using a group and a subgroup will create the table of 0 and 1's you are looking for. My interpretation of what you might be looking for is to use :Protein and :Sample as either the Group and Subgroup, respectively, or vice versa.
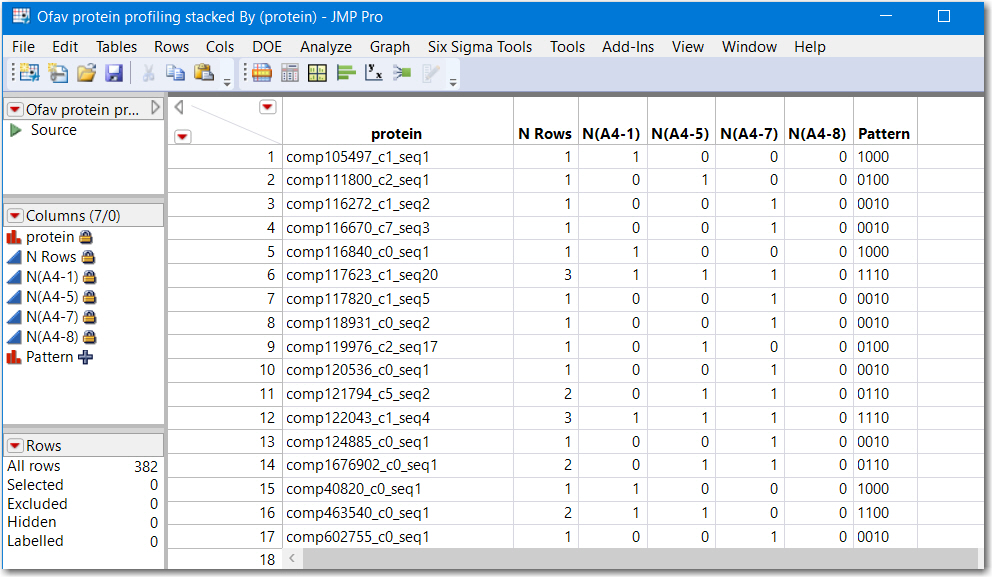
Below is a screenshot of using Summary with :Protein as Group and :Same as Subgroup. The JSL follows the picture.
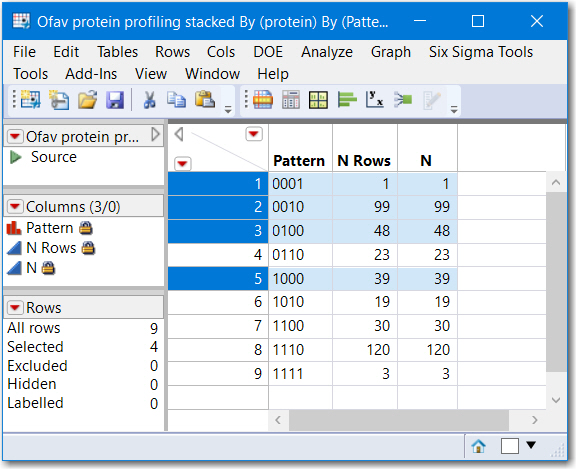
Note that rows where N Rows is greater than 1 represent proteins found in more than 1 sample. and rows where N Rows equals 1 is a proteinn unique to that sample. The column Pattern is a contatenation of the character 0 and 1's and is a numeric representation of "areas" of a venn diagram. Running another table summary by Pattern would reveal how many are shared.
Data Table( "Ofav protein profiling stacked" ) <<
Summary(
Group( :protein ),
N,
Subgroup( :sample ),
Freq( "None" ),
Weight( "None" )
)Summary by Pattern
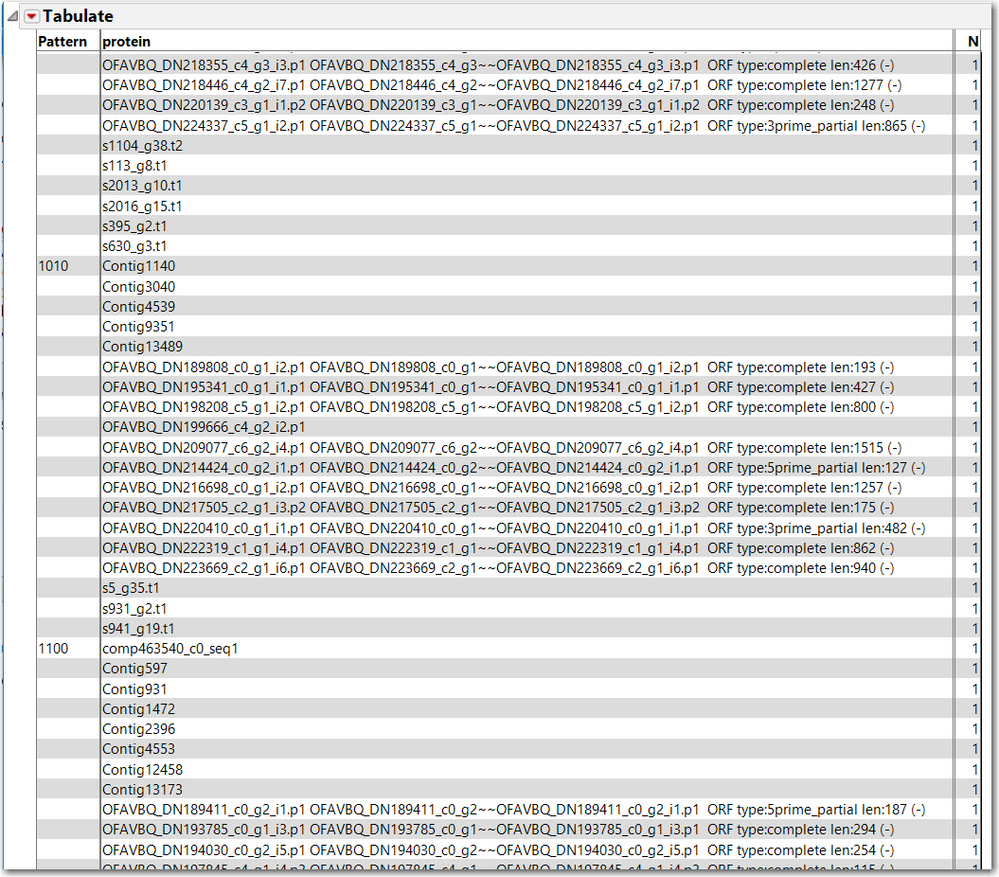
Also, Tabulate might be useful to provide the lists of common proteins for each pattern, not just N. This is the result of Tabulate, with Pattern and protein as grouping categories, only a small sample is captured below. This might not be exactly what you need, but hopefully provides some leads to your next steps.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Creating a Venn diagram based on protein frequency
I do not understand your data, especially how time and treatment should be factored into the analysis. I think Tables> Summary using a group and a subgroup will create the table of 0 and 1's you are looking for. My interpretation of what you might be looking for is to use :Protein and :Sample as either the Group and Subgroup, respectively, or vice versa.
Below is a screenshot of using Summary with :Protein as Group and :Same as Subgroup. The JSL follows the picture.
Note that rows where N Rows is greater than 1 represent proteins found in more than 1 sample. and rows where N Rows equals 1 is a proteinn unique to that sample. The column Pattern is a contatenation of the character 0 and 1's and is a numeric representation of "areas" of a venn diagram. Running another table summary by Pattern would reveal how many are shared.
Data Table( "Ofav protein profiling stacked" ) <<
Summary(
Group( :protein ),
N,
Subgroup( :sample ),
Freq( "None" ),
Weight( "None" )
)Summary by Pattern
Also, Tabulate might be useful to provide the lists of common proteins for each pattern, not just N. This is the result of Tabulate, with Pattern and protein as grouping categories, only a small sample is captured below. This might not be exactly what you need, but hopefully provides some leads to your next steps.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Creating a Venn diagram based on protein frequency
Brilliant! That is exactly what I wanted to do (despite not being able to properly elaborate it). I also forget about the tabulate function. The only thing I don't know how to do is create the "pattern" column, which will be very useful for making the Venn diagram. Did you just concatenate the prior columns or is there a specific "pattern" feature somewhere in the JMP package? Thanks so much for your help!
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Creating a Venn diagram based on protein frequency
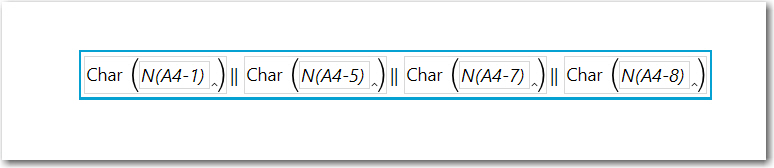
When wrote the other post, I used a column function (see below), however....
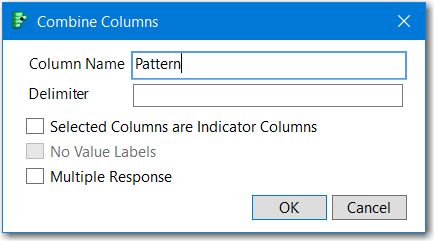
You can do this with Combine Columns. Highlight the columns representing Sample values. Then from the Main Menu, select Cols, scroll down and select Utilities> Combine Columns and the the dialog below will appear, remove the default comma delimiter, uncheck Multiple Response and name the column Pattern, OK and JMP will create the column to the left of the first Sample column..
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: Creating a Venn diagram based on protein frequency
Recommended Articles
- © 2026 JMP Statistical Discovery LLC. All Rights Reserved.
- Terms of Use
- Privacy Statement
- Contact Us