You might want to use logistic regression, where the disease class is Y and Assay is X. I am going to leave the labels 1-4 as they are, but I am going to tell JMP that the order is 2, 3, 4, and 1, because it assumes that the last level (1 now) is the target class (disease). Here is what it looks like when you regress Y versus X separately:
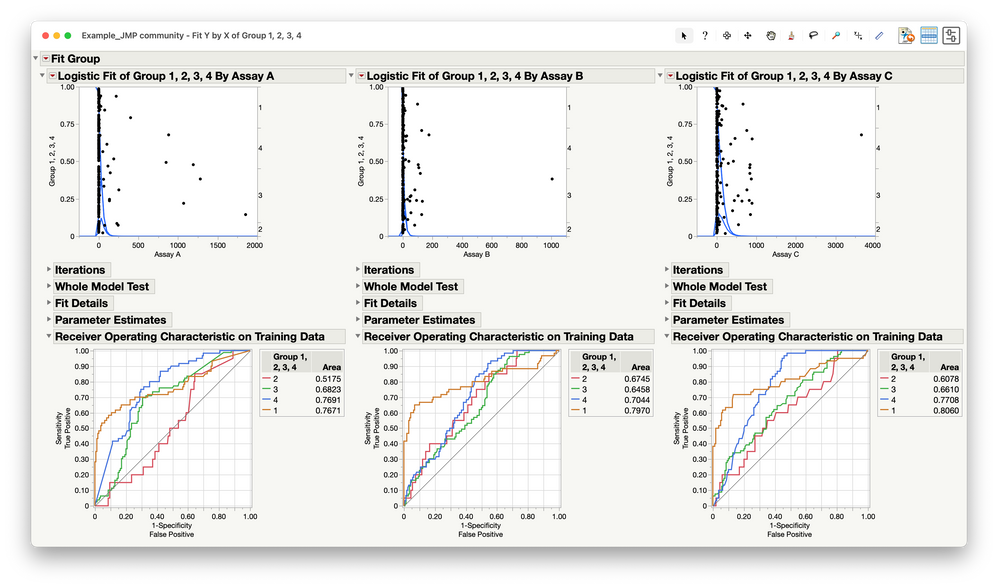
The ROC Curve is a diagnostic tool to assess discrimination (sensitivity versus 1-specificity) that might help you. It might also be helpful to use all three assays simultaneously as predictors to see if one is better. Here is nominal logistic regression that way:
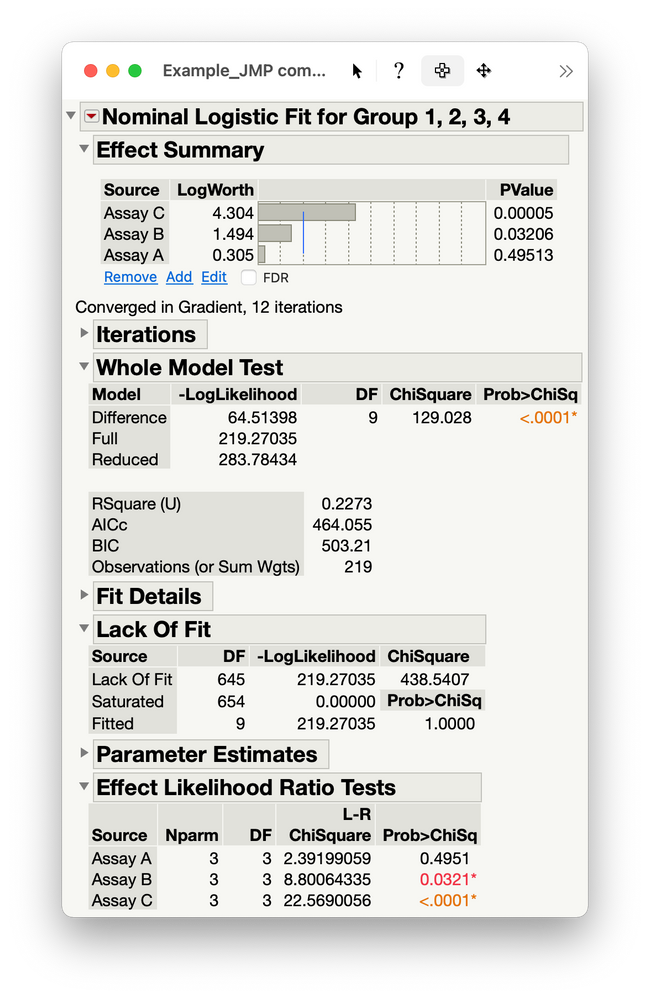
So it appears that Assay C does better than the other two assays. Now what is the best model? Is the relationship linear? Here is the fit and test of the cubic model:
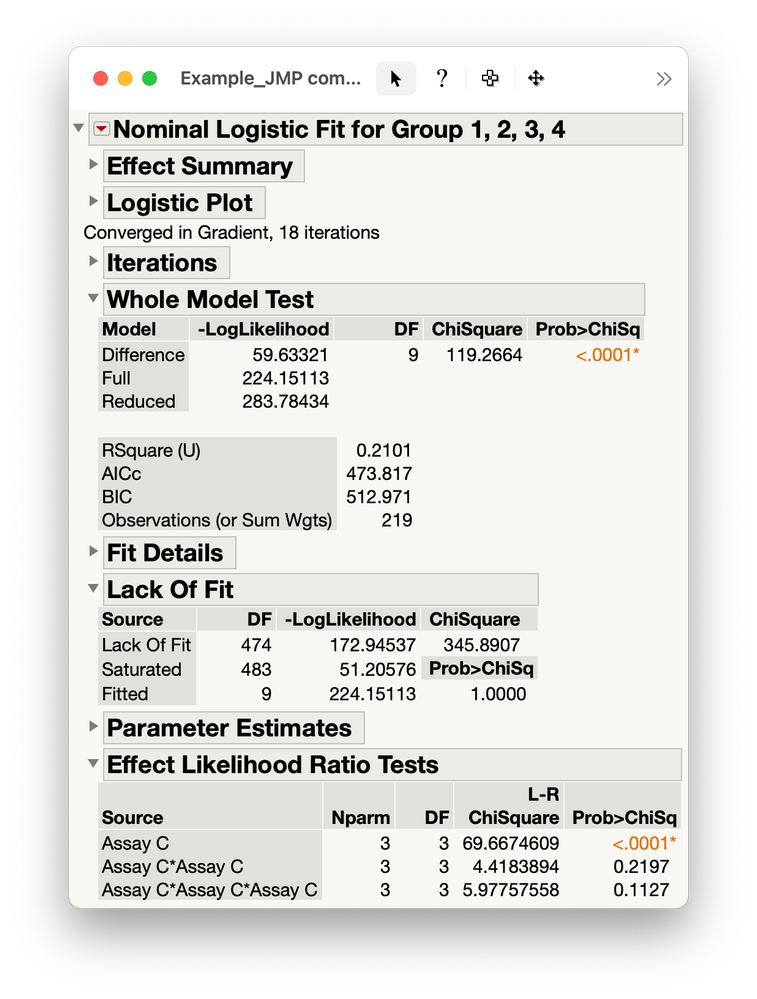
It seems that a linear predictor is sufficient.
What do the other classes (2-4) represent? Are there really 3 normal classes and 1 abnormal class? Also, is this study exploratory or confirmatory?
I attached your JMP data table after I saved scripts to repeat the analysis that I show here