- Subscribe to RSS Feed
- Mark as New
- Mark as Read
- Bookmark
- Subscribe
- Printer Friendly Page
- Report Inappropriate Content
JMP Add-Ins
Download and share JMP add-ins- JMP User Community
- :
- File Exchange
- :
- JMP Add-Ins
- :
- Rank Based Inverse Normal Transformation (INT) for Quantitative Trait Data
This JMP Add-in is to transform non-normal quantitative trait data typically analyzed in Genome Wide Association Studies (GWAS). The method implemented is rank-based inverse normal transformation (INT).
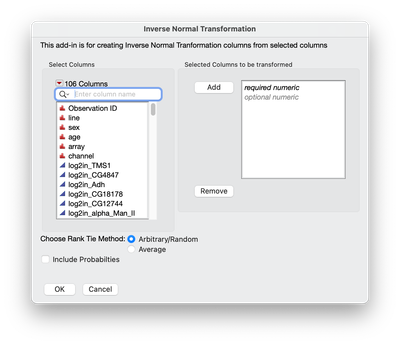
A simple interface is create to choose the traits to be transformed and then clicking OK to create new transformed columns (see screen shot below).
The output is two sets of columns: 1) _INT as a suffix. These are the transformed trait values 2) _p as a suffix. These are the probability values of the transformed columns.
There is a paper that discusses INT and its different forms. It also can be used to provide guidance.
McCaw ZR, Lane JM, Saxena R, Redline S, Lin X. Operating characteristics of the rank-based inverse normal transformation for quantitative trait analysis in genome-wide association studies. Biometrics. 020;76:1262–1272.
https://doi.org/10.1111/biom.13214
If there is interest, additional methods could be incorporated and the window can be updated to allow choosing these methods.
UPDATE: Version 2 of the add-in includes the below:
- Added option to include or not include probabilities
- Added an option to choose between Arbitrary or Average when there is a tie. Tie method chosen will write a note to the data table as to which method was used.
UPDATE 2: Version 3 of the add-in includes the below:
- Corrected a bug where total number of rows in data was counted (JSL function N Row()). The function for calculating rank INT should have used the number of rows with non-missing data (JSL Function Number()).
This add-in was created on, for and tested in JMP/JMP Pro 17 and JMP/JMP Pro 18
I needed help doing this analysis in JMP and Chris and Di, and perhaps others, were super responsive in getting this done.
Annabelle Rodriguez-Oquendo, MD
UConn Health, Farmington CT
Thank you for such a useful add-in. I have two questions:
1. when I used this add-in to transform the data it adds two columns to the dataset. One with "_INT" at the end of the variable name and another one with "_p". What is the second one ending with _p?
2. The paper that you have cited (McCaw et al., 2020) introduces two methods for inverse normal transformation; direct (D) and indirect (I) and then combine them to an omnibus test (O). I assume you only referred to the paper for more guidance for transformation. But in case the column is somehow related to one these three tests I appreciate if you can clarify which of them is used in the add-in.
Soheil Shapouri,
University of Georgia
- Mark as Read
- Mark as New
- Bookmark
- Get Direct Link
- Report Inappropriate Content
HI @SoheilShapouri,
_INT columns are the transformed columns, _p columns are the probability values.
Yes, the paper was more for reference as to the reasoning for the add-in and guidance. What was implemented was item 1. in the paper, the Rank Based Inverse Normal Transformation.
I will update the description to be more clear. Thanks for your questions and comments
Best,
Thank you, Chris, for custom creating these add-ins for me. I really appreciate you doing that when I needed them.
Recommended Articles
- © 2026 JMP Statistical Discovery LLC. All Rights Reserved.
- Terms of Use
- Privacy Statement
- Contact Us