A SMILES string is a textual representation of a molecular structure:
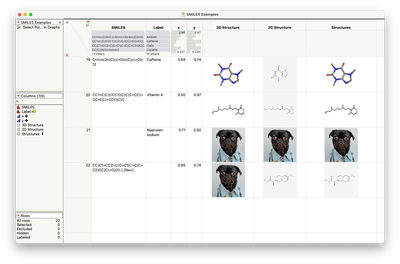
After assigning column roles:
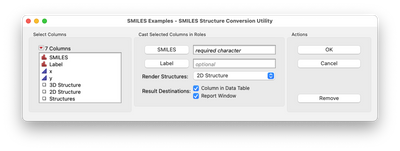
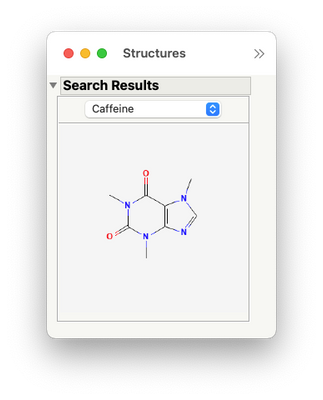
this add-in allows you to see a picture of the chemical structure(s) associated with any selected row(s) in the table (no matter how those rows were selected). The add-in has options to store the structures in a report window,
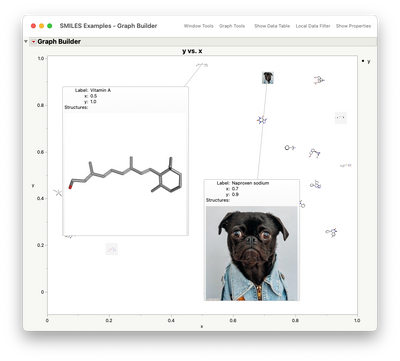
or in the data table itself for use as markers or in hover windows (perplexed puppies are automatically substituted when a structure can't be found).
The add-in was originally written by Ian Cox, Graeme Robb of Astra Zeneca, and Russ Wolfinger of JMP, so thanks are due to them. The Add-in is currently maintained by Mike Anderson of JMP (questions, feedback, or feature ideas should go to him).
V2 Released 08Nov2020
- Uses the NIH PubChem REST API to pull 2D or 3D structures for a SMILES reference.
- Updated code base to take advantage of Expression Columns and other new JSL structures.
- Added perplexed puppies
- Added support for MacOS