- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Discussions
Solve problems, and share tips and tricks with other JMP users.- JMP User Community
- :
- Discussions
- :
- Re: How to address weak heredity in screening designs?
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
How to address weak heredity in screening designs?
I'm about to prepare a DoE for a novel process which involves up to 11 continuous factors while one of these is hard to change.
Since I'm trying to combine this with an effort to make the ideas behind DoE a bit more transparent to colleagues, I've come across a few questions, which can be summarized like this:
What's the best design approach when domain knowledge suggests the existence of two-pair interactions and possibly quadratic effects, while I cannot assume strong heredity?
I'm hesitant to create a custom design involving "all" terms (i.e., main effects, two-way interactions and quadratic effects) as it looks like a brute-force solution and contradictory to an efficient experimentation. I would rather prefer to do an initial screening design to (hopefully) ignore non-contributing factors and run a subsequent augmented design. My major concern is that an improper screening design may lead to the loss of important factors due to a missed interaction or quadratic term.
It seems that there are a few threads with similar questions but touching only little aspects of this, so I'm posting this as a new thread here.
In order to study the design and analysis methods, I've setup a test case which I'm attaching. I've created a definitive screening design for five factors, including a central point. For the sake of simplicity, I've ignored the possible existence of hard-to-change factors. The target variable Y is generated through a formula, where you'll see the relevant terms.
I tried several approaches to find the proper model (since I have a priori knowledge about the true function, I'm always biased) and I don't want to explain in all detail what I've done but a brief summary is:
- From a pure screening for main effects, I might drop factors X2, X3 and X4 and might not include them in a subsequent augmented design.
- By fitting a definitive screening design, I would be mislead to interactions that aren't there and also quadratic terms would be confused. I'd have to drop the assumptions of strong heredity for both interactions and quadratic terms.
While I understand where this is coming from (strong correlations between interactions and also quadratic terms within the given design), the question is, how to best deal with this case?
I understand that my target function is made up arbitrarily and hence, it might well be a fairly pathological case. I'm wondering how often this issue arises in practice?
Looking forward to your feedback.
Björn
Accepted Solutions
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: How to address weak heredity in screening designs?
Adding my thoughts to the excellent thoughts by @P_Bartelland @Mark_Bailey I suggest you use scientific method to pursue your investigation. That is, start with hypotheses, get data then modify, drop, add hypotheses and iterate. It is impossible to know, a priorí, the best design (if you knew this, you probably don't need the experiment). Advice is always situation dependent, so without a thorough understanding of your actual situation, I can only provide general advice. That is to start your investigation creating as large a design space as resources will allow (lots of factors at bold levels). Don't under-estimate the effect of noise! Have a strategy to handle all of the other factors not being explicitly manipulated in the experiment (e.g., repeats, replicates, blocks, split-plots). The larger the inference space the more likely your results will hold true in the future. You are looking for direction (which you can get with 2 levels) and which factors are most likely to move you to the best space quickly (linear effects). Subsequent experiments will focus on the design space with the best results. It is OK to confound effects in initial experiments. Confounding is an efficiency strategy. The key to using confounding efficiently is knowing what is confounded. This way, subsequent iterations can separate confounded effects. Do this; As an SME, predict the rank order the model effects up to 2nd order (all main effects and interaction effects). If the top of the list is all main effects, then you can use low resolution designs in your first iteration. You are looking at the relative importance among those main effects. As the 2nd order effects rise in your list, you will want to consider higher resolution designs.
In any case, design multiple options of experiments (easy to do this in JMP). Evaluate what each different design option will potentially get you, in terms of knowledge (e.g., what effects will be separated, which will be confounded and which won't be in the study). Contrast this with the resources required to get that knowledge. Pick one that meets your criteria of knowledge vs., resources. Keep in mind you will be iterating, so don't spend your entire budget on the first experiment.
Mark is entirely correct. The fractional factorial strategy uses the principles of Scarcity and Hierarchy. Heredity only comes into play when doing analysis and trying to assign which of the confounded effects are most likely active. Prediction of which of the confounded effects is most likely from the SME is the most powerful method. If prediction and heredity can't provide guidance, then you'll need more data.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: How to address weak heredity in screening designs?
All things considered I'm thinking a definitive screening design approach might be your 'best', and that's a relative term, option. If you are unwilling to go the optimal design approach I can't think of a better alternative...others please chime in. One statement you made though confuses me a bit:
"By fitting a definitive screening design, I would be mislead to interactions that aren't there and also quadratic terms would be confused. I'd have to drop the assumptions of strong heredity for both interactions and quadratic terms."
Why would a DSD be prone to indicating interactions that aren't there? In fact, I think it's the other way around...you may never find active interactions because of the relatively rare instance where a two factor interaction is active, but the main effects are not. My experience says this situation doesn't happen often. As long as you use the Fit DSD workflow you can guard against, to some degree, this issue.
Plus...think about experimenting sequentially. Try the DSD. Hopefully you aren't in a place where you are forced to solve the problem with one and only one experiment. Again, rare is the occasion, in my experience, where this happens. Especially with a less than well understood system of study. See what you learn. Then based on what you learn, confirmation runs, more experimentation in the optimal DOE space, are two ways you might go.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: How to address weak heredity in screening designs?
Thanks for your reply! Sorry for not coming back to you ealier as I didn't have access to the community.
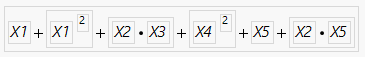
Let me clarify my initial statment which gave you some concern. The function that the target quantity follows is this:
As you can see, the interaction x2*x3 contributes but not the two main factors x2 and x3. Running the DSD fit does not reveal x2*x3, neither does if propose any importance for x2 or x3. That's essentially what I wanted to express: I might drop the idea of following up on x2 and/or x3 in a subsequent DoE and might miss the x2*x3 interaction.
I agree with you that in practice we hardly would find the case of having the interaction without any contribution of main effects. I understand that the modeling approach here is essentially fitting a Taylor series to the "true function". On the other hand, I was worried a bit by the fact that, for example, many interdependencies/relations in physics follow a formula were one quantity is a product of powers (negative and positive) of the other quantities. Again, I think it's as you said: it's more of an academic question but a practical one.
For my specific application, I had considered running a DSD but there are two caveats:
- I have a hard-to-change factor
- Two variables have a linear interdependency: both cannot take their minimum at the same time, which I would avoid through a linear boundary condition.
I would then judge the optimality of my design by looking at the correlation matrix and if necessary, add model terms if the remaining correlations are still to strong.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: How to address weak heredity in screening designs?
In addition to @P_Bartell's advice, I want to add two considerations. Screening designs work best when the situation exhibits the key screening principles of effect sparsity and hierarchy. You expect most of the factors in the design to be inactive. Is that the case here?
I usually recommend not separating factors into separate designs unless there is some constraint. Two factors with interacting effects will not exhibit the interaction if they are designed separately. You treat the factors held out as in a one-factor-at-a-time experiment. If you expect large effects, you can use a DSD with 23 runs. Model heredity only comes into play during the analysis.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: How to address weak heredity in screening designs?
Thank you for your thoughts!
In fact, from domain knowledge, we actually expect quite a number of active factors. From the 9 factors under consideration (I stated 11 in my intial post, but it's 9), I think that 6 or more could be active. I would agree that separation of factors wouldn't be a good idea especially since we believe to have interactions.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: How to address weak heredity in screening designs?
Adding my thoughts to the excellent thoughts by @P_Bartelland @Mark_Bailey I suggest you use scientific method to pursue your investigation. That is, start with hypotheses, get data then modify, drop, add hypotheses and iterate. It is impossible to know, a priorí, the best design (if you knew this, you probably don't need the experiment). Advice is always situation dependent, so without a thorough understanding of your actual situation, I can only provide general advice. That is to start your investigation creating as large a design space as resources will allow (lots of factors at bold levels). Don't under-estimate the effect of noise! Have a strategy to handle all of the other factors not being explicitly manipulated in the experiment (e.g., repeats, replicates, blocks, split-plots). The larger the inference space the more likely your results will hold true in the future. You are looking for direction (which you can get with 2 levels) and which factors are most likely to move you to the best space quickly (linear effects). Subsequent experiments will focus on the design space with the best results. It is OK to confound effects in initial experiments. Confounding is an efficiency strategy. The key to using confounding efficiently is knowing what is confounded. This way, subsequent iterations can separate confounded effects. Do this; As an SME, predict the rank order the model effects up to 2nd order (all main effects and interaction effects). If the top of the list is all main effects, then you can use low resolution designs in your first iteration. You are looking at the relative importance among those main effects. As the 2nd order effects rise in your list, you will want to consider higher resolution designs.
In any case, design multiple options of experiments (easy to do this in JMP). Evaluate what each different design option will potentially get you, in terms of knowledge (e.g., what effects will be separated, which will be confounded and which won't be in the study). Contrast this with the resources required to get that knowledge. Pick one that meets your criteria of knowledge vs., resources. Keep in mind you will be iterating, so don't spend your entire budget on the first experiment.
Mark is entirely correct. The fractional factorial strategy uses the principles of Scarcity and Hierarchy. Heredity only comes into play when doing analysis and trying to assign which of the confounded effects are most likely active. Prediction of which of the confounded effects is most likely from the SME is the most powerful method. If prediction and heredity can't provide guidance, then you'll need more data.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: How to address weak heredity in screening designs?
Thanks for your excellent feedback! I will start with a custom design including as many factors as possible taking the boundary conditions into account.
Technically speaking, the scientifc method is what I am accustomed to but the challenge for the method sometimes lies in the acceptance of the method. Especially for DoE, performing seemingly wild and random factor settings with many factors may confuse people not acquainted with the reasoning behind DoE. Hence, if one has to toss (or refine - to use a more pleasing wording) one's hypotheses too often, the culprit might be seen in the method and not necessarily in the fact that some problems are simpliy trough to solve (for whatever reasons like missing factors, noise etc. etc.).
For my specific issue, I'm planning to set the factors to "reasonably" extreme min and maximum values for screening, hoping to quickly eliminate a few factors and then refine with an augmented or even new design, depending on the results and trends.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: How to address weak heredity in screening designs?
I thought I give a little follow up on the topic. I've created a custom design including main and quadratic effects for 9 factors. As I've said, there's a factor which is difficult to change (X1) and two factors (X2 and X3) are subject to a linear boundary condition (X2+X3 > const.).
This is the correlation matrix resulting from the custom designer:
The analysis after collecting the data showed that up to 6 out of 9 factors are active. It also happened that X2 itself isn't active but from physical considerations it should to be as its nature is the same as X3. Hence, I've tested for X2^2 and this is a significant contribution.
I hope my conclusion isn't jumping too far but it appears to me that the principles of sparsity and hierarchy are violated here.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: How to address weak heredity in screening designs?
I don't see the data table attached, so it is impossible to analyze the data. How did the residuals look? Is there any collinearity between X2 and X2^2 (I notice X2 is not in your matrix?)? Did you go too bold on the X2 level setting? Do they all have practical significance? It is easy to have statistical significance, just minimize the factors that constitute the MSE.
The principles are just that. They are not absolutes. They are for guidance. I will still suggest that 6 of all of the possible factors (much greater than the 9 you experimented on) is a small subset.
Recall F=MA...neither M or A are significant (both first order effects).
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Get Direct Link
- Report Inappropriate Content
Re: How to address weak heredity in screening designs?
Hi,
I hadn't attached the data table, only the screenshot of the correlation matrix of the experimental design. X2 is in the design. As JMP omits labels if the labeling becomes too dense, it's simply not labeled. X2 would be the second row or column, respectively. The design included the main effects as well as all quadratic effects. The correlation of X2 and X2^2 is 0.1.
The factors levels are reasonable based on our experience.
Overall, I'm wondering if the underlying physical model is more complex, over the given variable range, for being well fitted with a second order taylor polynomial (thinking of terms like radius of convergence etc. etc. - guess that would be a conversation for a conference and not so much for a forum discussion).
Recommended Articles
- © 2026 JMP Statistical Discovery LLC. All Rights Reserved.
- Terms of Use
- Privacy Statement
- Contact Us