- JMP User Community
- :
- File Exchange
- :
- JMP Add-Ins
- :
- XGBoost Add-In for JMP Pro
- Subscribe to RSS Feed
- Mark as New
- Mark as Read
- Bookmark
- Subscribe
- Printer Friendly Page
- Report Inappropriate Content
New for JMP 17 Pro: An Autotune option makes it even easier to automatically tune hyperparameters using a Fast Flexible Filling design.
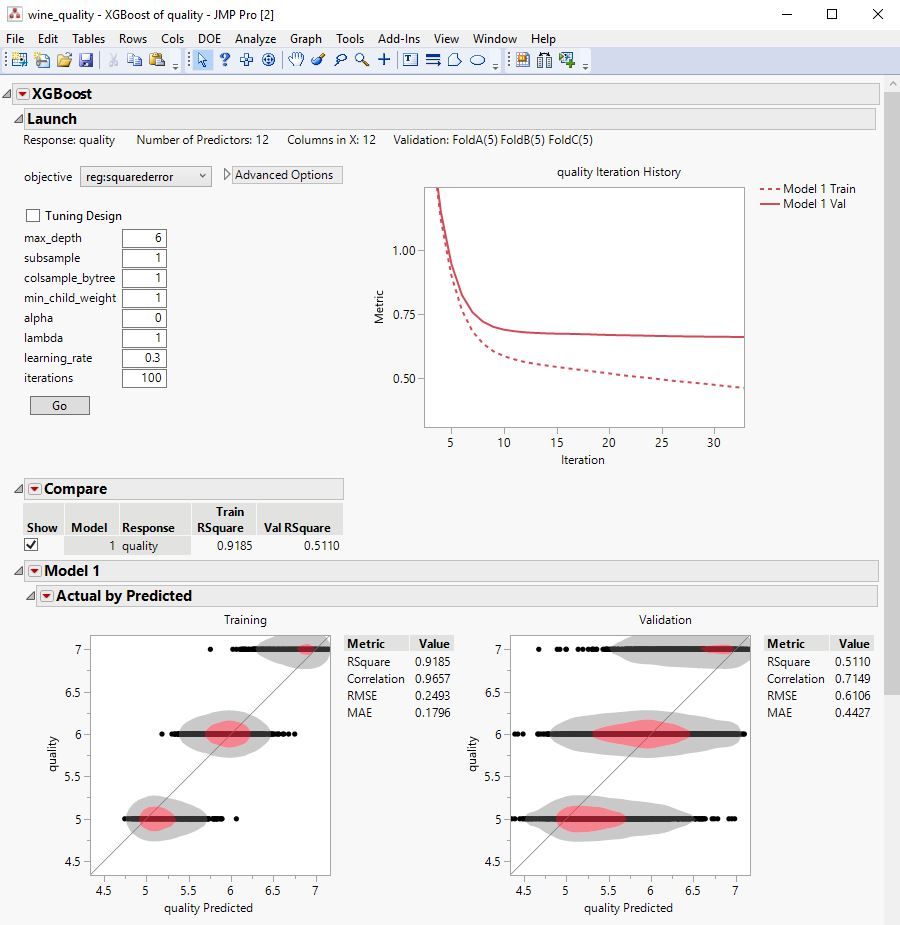
The XGBoost add-in for JMP Pro provides a point-and-click interface to the popular XGBoost open-source library for predictive modeling with extreme gradient boosted trees. Value-added functionality of this add-in includes:
• Repeated k-fold cross validation with out-of-fold predictions, plus a separate routine to create optimized k-fold validation columns, optionally stratified or grouped.
• Ability to fit multiple Y responses in one run.
• Automated parameter search via JMP Design of Experiments (DOE) Fast Flexible Filling Design
• Interactive graphical and statistical outputs
• Model comparison interface
• Profiling Export of JMP Scripting Language (JSL) and Python code for reproducibility
Click the link above to download the add-in, then drag onto JMP Pro 15 or higher to install. The most recent features and improvements are available with the most recent version JMP Pro, including early adopter versions.
See the attached .pdf for details and examples, which can be found in the attached .zip file (along with a journal and other examples used in this tutorial).
Related Materials
Video Tutorial with Downloadable Journal: XGBoost Add-In for JMP Pro (2020-US-45MP-540)
Add-In for JMP Genomics:
Add-In from @Franck_R : Machine Learning with XGBoost in a JMP Addin (JMP app + python)
XGBoosters,
Wanted to let you know there have been a few fixes and updates to XGBoost in JMP Pro 17.2, which was just recently released. These include updated default parameters, SHAP values, and scripting Advanced Options. The same add-in works since changes are on the JMP side.
In addition, JMP 18 Early Adopter 5 is due to release soon. It includes a brand new Torch Deep Learning add-in that has an interface very similar to that of XGBoost and offers many kinds of predictive models never before available in JMP Pro (image, text, and deep tabular neural nets).
If you like XGBoost, you're gonna love Torch Deep Learning. Please sign up for JMP 18 Pro EA5 at https://jmp.com/earlyadopter , give it a try, and send feedback.
Hi @russ_wolfinger ,
I'm running JMP Pro 17.0.0 on a Windows 11 64-bit machine (work laptop), 12th gen Intel i5 core.
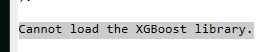
Have been using XGBoost since the EA version several iterations ago. Haven't had much problems with it until recently when I tried to run XGBoost. Unfortunately, I get the following error.
I've tried unregistering and re-installing the add-in, but it's still not working. Is there any known issue with this or how to work around it? I've called it in to JMP tech support as well.
I don't know if it matters, but when I go to the com.jmp.xgboost/lib directory, there is only one directory there called 15, which has 7 files in it. Should there be one for 17?
As an aside, I'm also running JMP Pro 18.0.0 EA 5 on my personal machine and can run XGBoost just fine -- along with the new Torch Deep Learning (which is really cool!). On my personal machine, the lib add-in directory has two directories, 15 and 17. Maybe it's related to this?
Thanks!,
DS
@SDF1 Strange, yes, that 17 subfolder should definitely be there and would explain the error message. I'm not sure at this time why it would have disappeared. A few ideas to fix: 1. Completely delete the com.jmp.xgboost folder then re-install the add-in. Verify the 17 subfolder gets created. 2. Copy and paste the 17 subfolder from your other machine. 3. XGBoost.jmpaddin is actually a zip file. Unzip it into a temporary folder, then copy and paste the 17 subfolder into the JMP add-in location.
Hi @russ_wolfinger ,
Thanks for the quick feedback. Not sure what happened, but I followed the suggestion to unregister the add-in, delete the directory and re-install after downloading the latest version. Worked just fine, thanks! Now I can run XGBoost again!
Thanks,
DS
Dear Dr. Wolfinger,
Regarding your post about testing the newest version of the JMP earlyadopter, can I indicate you as a reference? Once upon a time I participated in this testing (JMP 17), and if you do not mind, I would check fixed XGBoost in this up-to-date version (18). Thank you!
@Nazarkovsky Certainly! Really appreciate all of the great feedback you have provided over the years, and it would be very nice if you could check out the latest XGBoost. I've been spending most of my time this year on the Torch Deep Learning add-in and since its interface is very similar to XGBoost's, have found a few opportunities to improve both.
Hi @russ_wolfinger ,
I don't know if this has been corrected for JMP Pro 17.2, but I just noticed a strange omission of the Profiler when re-running a saved XGBoost script from the data table.
So, after I've run an XGBoost fit and open up the profiler, if I then save the script to the data table and try to re-run it, it doesn't automatically bring up the Profiler like in the other fit platforms. It would be great if that could be updated if it hasn't already. For JMP Pro 17.0.0 at least, it doesn't pull up the Profiler from a saved script. Or, is there some setting I'm not aware of to get XGBoost to do add it to a saved script?
Thanks!,
DS
@SDF1 Thanks a lot for reporting the Profiler script not running. This is a bug and I've just fixed it (also applies to other post-fit commands like Save Predicteds). The next JMP 18 early adopter release (EA7) should have the fix. Unfortunately I don't see any workarounds with the current version as the XGBoost parser was effectively ignoring all post-fit scripting commands.
You get double extra credit because the same problem was in the Torch Deep Learning code and should also be fixed in EA7.
hi Russ,
I am using JMP 16 Pro. I loved your work.
I am having trouble though in getting any Tree Details. The white space expands but nothing comes up.
Do you know the solution ? I think I downloaded the addin a few weeks back, has there been an update since then ?
thx.
hi Russ,
Could you pls address my prior question. I am unable to get any trees from your XGboost - Pro 16 and 18EA both did not deliver.
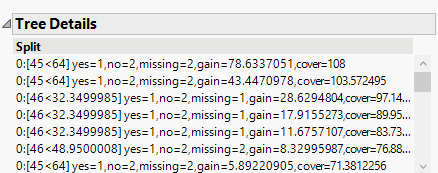
Hi @altug_bayram Apologies for the delay. When clicking a model red triangle > Tree Details, you should see a new outline box at the end of the output. The attached image is from the Press Banding example that comes with the add-in. Does this example work for you?
Another related idea is to click red triangle > Save Prediction Formula and then examine the formula of the newly created column to see the logic of the prediction. If still not working, please feel free to email data and model to russ.wolfinger@jmp.com and I can take a look.
This outline is coming out but is difficult to translate as parameters are replaced by indexes.
Do they refer to the ordering under Importances or something else ?
Isn't it possible to get regular tree diagrams (as would be produced by Bootstrap Forest for instance ) ?
Also, my addin is not under regular JMP directory structure but rather under an independent windows folder. Should I remove/reinstall ?
Also, having confirmed that I see the outline (but difficult to translate) , is there any advantage to gain by using 18 Pro EA as opposed to 16 Pro ?
thx.
hi...
I cannot send you any data or model.
Do you have any samples that show how to interpret the tree details. As these are not in the standard tree diagrams, it is difficult to understand w/o some sort of example.
Hi @altug_bayram The add-in parses these strings in C++ code to extract the tree splitting rules and variable importance values. I think the first number is the 0-based index of the predictor X variable in the order specified for the model. I'm not aware of any official doc about them--we deduced parsing rules after studying them. The add-in currently offers no way to plot the trees as there are typically hundreds of them in a k-fold fit, but as I mentioned, the logic is shown in the prediction formula which is presented similar to a tree if you save the prediction formula then click on it Also, the following R package looks like it plots the trees in case you want to try it: https://rdrr.io/cran/xgboost/man/xgb.plot.tree.html
Add-ins are installed in a folder separate from the JMP installation location to ensure you have write permissions.
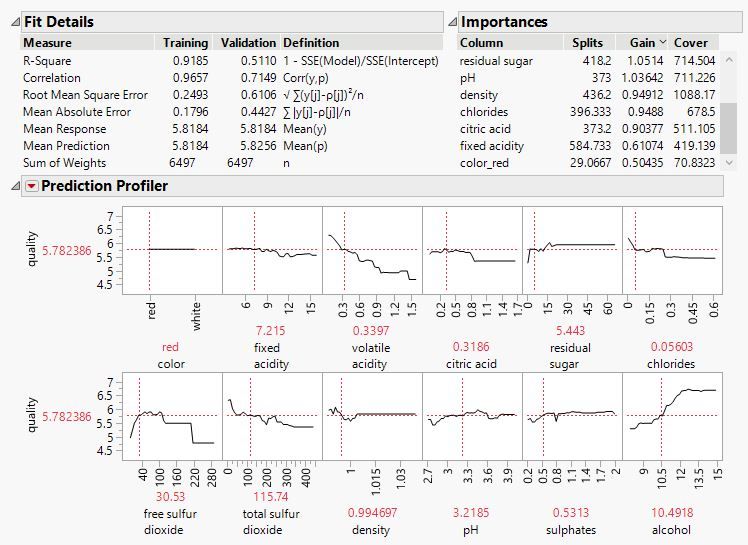
There have been a lot of improvements to the add-in since version 16--please scroll back through this thread to see posts about them. Assuming one of your goals is to interpret what the model is doing, some very useful outputs in this regard are Shapley values. They break down each row's prediction into components attributable to each variable. It can be very informative to plot Shapley values versus the variable they correspond to. You can obtain Shapley values by clicking a model red triangle > Save SHAPs.
Hello Mr russ_wolfinger ( @russ_wolfinger )! Is there a version of this wonderful add-in that is not just for JMP PRO?
Hi @rubenslana Alas, no, but we happen to think JMP Pro is worth it so please request it
- « Previous
-
- 1
- 2
- Next »
Recommended Articles
- © 2024 JMP Statistical Discovery LLC. All Rights Reserved.
- Terms of Use
- Privacy Statement
- About JMP
- JMP Software
- JMP User Community
- Contact