-Hi, I'm Jerry Fish I'm a technical engineer with JMP.
I cover several Midwestern states in the US.
I'm joined today by one of my colleagues Scott Allen,
who's also a technical engineer for JMP
and he supports several other Midwestern states.
Hi, Scott.
-Hey, Jerry, good morning.
-Good morning.
Today we want to talk about a variety,
a relatively new way to analyze data, specifically from milling operations,
where we want to learn about milling to help with a scale- up milling process.
-W e both have a strong interest in process optimization using DOE
and in modeling response curves
using the Functional Data Explorer in JUMP Pro,
and we wanted to bring those together for today's presentation.
And one of the first examples I saw doing this sort of analysis
was the milling DOE that's in the sample data library,
where the goal is to optimize an average particle size.
So as we were talking about possible topics for today,
we thought it would be interesting to see if we could extend that
instead of optimizing that just the particle size,
could we actually optimize the particle size distribution response curve?
-So, milling has many different applications.
You'll find it in anything from mining, food processing,
making toner in the printing industry, making pharmaceutical powders.
At the most basic level, a milling process is used
when we want to reduce the particle size of a certain substance
and produce uniform particle shapes and size distribution
of a starting material.
Often some type of grinding medium is added to accelerate the process
or to control the size and shape distribution
of the resulting particles.
In each application the desire is to get the right particle size,
say the median or the mean particle size,
with a controlled predictable particle size distribution.
In the scenario we discussed today,
we have an existing manufacturing milling operation
that produces a pharmaceutical powder.
It has good performance today,
creating the right medium particle size and a narrow particle size distribution.
A full disclosure this scenario and the resulting data are invented.
Scott and I didn't have access to non- confidential data
to present in this paper.
However, even though the data are fabricated,
the techniques that we're about to show are applicable for real world problems.
The picture on the left shows a typical agitated ball mill.
The material to be milled, enters at the top
and continuously flows into the vessel where an agitator rotates
the material and the grinding medium to affect particle size.
The resulting particles are then vacuumed out of the vessel
as they're milled.
Management has said they need to increase the production output,
something we're all, I'm sure, familiar with.
They are considering building a new milling line,
but before investing all that capital, they'd like to investigate,
can we simply increase the throughput of our existing equipment?
So manufacturing made some attempts at doing that,
they adjusted their process,
and while they can affect the median particle size,
the new output has odd particle size distributions.
So manufacturing came back to R& D where Scott and I are,
and asked us to go to the pilot lab
and see if there's any combination of settings that might improve throughput.
Scott, what parameters did we look at?
-In this case,
we're going to use six different factors for a DEO.
There's going to be four continuous factors
so agitation speed, the flow rate of carrier gas,
the media loading as a percentage of the total,
and then the temperature of the system.
And then there's two categorical factors,
the media type and maybe the mesh size of a pre screen process.
And so determining those factors
is fairly straightforward these are known to affect particle size distributions
and things like that, but the response is still a challenge.
And in this case, I'm not sure how you would actually go bout doing this
if you couldn't model the response curve like we're going to do.
So Jerry, in your experience, how would you have done this before?
-S o this next slide shows typical particle size distribution
or particle size density plot.
And we've got a plotted as percent per micron versus size.
But you can think of this as just a particle count
on the y- axis or a mass distribution,
it's just a histogram representing the distribution of particles.
What you're seeing on your screen,
this might be a good particle size distribution,
has a nice narrow shape with a peak
at the desired median particle size.
But how do we characterize this distribution
if we want to do a test to adjust it?
Well, we might characterize the location with the mean, median, mode
of the distribution.
And we might characterize the width via a standard deviation
or maybe a width at half peak height,
those are typical ways we might measure that.
But when manufacturing tried to turn various production knobs
in their process to speed up the throughput,
they saw varying degrees of asymmetry in the distribution.
Maybe this was due to incomplete milling of the pharmaceutical material,
or perhaps there were temperature effects that caused particles to agglomerate,
we don't really know.
But now the half width
isn't really representing the shape of that new curve.
So we might turn to calculating maybe percentiles
along the width of the curve,
maybe the 10th percentile of particles that fall below a certain point,
20% below this point, 90% below this point, and so forth.
But it gets even tougher when we're trying to describe something like this shape
where there are two very pronounced peaks, or this shape,
which I tend to call a haystack where it's very broad, doesn't have tails.
What do we do with that?
So this parameterization technique doesn't seem to be the best way
to approach the problem.
Scott, I know we have to do some experimentation,
but how are we going to approach this in today's analysis?
-So that's what we're going to do.
So we're going to use the entire shape,
our response in this case is going to be that curve.
So we're not going to try to co-optimize all those different parameters
that you talked about.
We could co optimize two, three, four different parameters,
but instead we're going to use that entire curve as our response.
We're going to use all the data and then our target
is going to be some hypothetical curve that we want to achieve.
So once again, we're not going to try to target
all the different parameters in that curve,
we're going to try to match the shapes.
So we want to have our experimental shape match the shape of our target.
And so that's how we're going to get started with the analysis today
and we'll take you through the workflow of how we would do that.
So let me go and get into JMP.
Oops, there we go.
So we first see here is the DOE that we ran.
So we ran a definitive screening design with those six factors,
although you could use any design that you wanted.
And we've got 18 experiments in this case,
so here's all the factors and the factor settings that we used.
-That looks pretty standard to me, Scott, for the DOE that I've run in the past.
But you don't have a response column.
-W ell, that's one of the unique things about the response curve analysis is
in some cases you set it up a little bit differently and how you do the analysis.
So we don't have a response column
and we're not going to optimize just a single value.
Instead, our responses are in this other table.
So in this case, we've got a very tall data set
with the x- axis is our size and the Y value is our percent per micron
and this is what we're going to plot and optimize.
But we do need to get our DOE factors in there.
So to do that, we just took a little bit of a shortcut
and we did a virtual join between these two tables.
So in our design here, our run number is the link ID.
And then we've got the run number here and this is the link reference,
and this lets us bring in all of those DOE factors.
So these are all here in the table, but they're just virtually joined
and that helps us keep our response table nice and clean.
So if there's any modifications, we don't have to worry about copying
and pasting or adjusting all those DOE factors.
So that's how we set up our table,
we've got our DOE factors in this table and our DOE responses occurs in this.
And as you can see, we've got all of our 18 runs here.
And before we start the analysis, let's take a look at those curves.
So I just plotted all those curves in Graph Builder,
and so we've got our target curve here.
So this is a hypothetical target curve that we want to achieve,
it's going to be experiment number zero.
And then we've got experiments one through 18
and the response curves for each of those.
-So you've run 18 experiments and not a single one of those
looks exactly like that target.
What do we do with that?
-That's a good observation.
And in this case, what we can see are some different features between these,
so definitely some are more broad.
I like how you called it, that haystack here.
Some are more narrow, maybe with smaller shoulders here.
We do see that the peak shifts a little bit in some of these,
here's the peak, it's shifting left and right.
And so hopefully we can find some settings and those factors
that will use the best of all of these give us something that's narrow
without a shoulder or bimodal peak.
But to do that, we need to go
into the Functional Data Explorer.
This is a traditional DOE, we would go up to analyze
and we would go to fit model, potentially.
In this case, we're going to go down here
to specialized modeling and go to Functional Data Explorer.
And so when we launch this, we need to add our Y values,
which were the percent per micron.
The X values was our micron size.
We need to identify each of those functions
with the run number.
And then we're going to add all those DOE factors
as supplementary information.
So I'm just going to take all of my DOE factors
and add them as supplementary information.
Now we'll click okay.
So when you launch the Functional Data Explorer,
this is what you get first.
And this is just a data processing window.
And what we're doing is just taking a quick look at
all of our data.
And so in this initial data plot, we just have all of our curves overlaid,
and you can see our green target curve hiding in there.
So this just shows us how all of our data are lining up.
Over here on the right, we have a different set of functions
to help clean up the data, process it.
And one of the really nice things about this platform
is you don't have to do that data processing in the data table.
So if you needed to remove zeros or do some sort of adjustment here,
standardized the range, things like that,
you can do all of that over here in the Clean up.
In our case, our data is pretty clean,
so we don't need to do any data processing.
But what we do need to do is take this green curve,
our target curve, out of the analysis.
So this is the target, this is what we're going to try to match.
And so we don't want it to be part of the modeling analysis.
So to take that curve out, we go over here to the target function
and we click load.
And I'm going to select that zero curve, click okay.
And now it's gone.
So now we're not going to include that in our models.
And so we can scroll down here and just see how each
of our individual experiments are plotted.
So now that our data is nice and cleaned up,
we can go on to the analysis.
So to do the modeling, we go up to the red triangle
and there are several different models that we can choose.
And in a typical workflow,
at the beginning, you might not know which is the best model to use,
whether you're going to use a B-spline or a P-spline or something else.
In this case, in the interest of time, we've done all of that already.
And we know that the P-spline gives us a pretty good model.
So we're going to go ahead and fit that model.
So I select P-spline and now JMP is creating the models.
And what we'll see is,
the first thing we'll is something similar to that initial data window over here.
So all of our curves are still plotted and overlaid.
But now we've got this red line and this red line is representing the mean
so the mean curve of all of those different curves.
And so we can also scroll down below
and we see each of our individual experiments
also with a line of fit.
And this is the first indication that you can get about
how well this model is fitting.
So if you're getting all these red lines overlaying your experimental data,
then you're probably on the right track.
If there was a lot of deviation
then you might consider doing a different model.
Other thing you'll notice over here is there's different fitting functions
that the spline model is using.
In this case, there were two that JMPs that are pretty good.
So this linear model and the step function model
and by default all the analysis down below
is going to use the model that had the lowest BIC value.
So in this case, all the analysis is using this linear model.
But if you wanted to use a different one you just select it
and I don't know if you can see it easily, but this one's highlighted now
or you go to the linear or you can just click on the background
and it'll go to the default.
And so that's the modeling side of it.
But we need to check how well this model is fitting.
And so to do that, we just go down here to the window that has a functional PCA.
So this is the functional principle component analysis.
This looks a little complicated, but what we want to do
is really start to take a look at how well this model has been created.
And so what we want to do is look at this mean curve here.
So this is the same mean curve that was calculated in the section above.
And what JMP has done is said, we're going to start with this mean curve
and then we're going to add a shape.
So we're going to add this function or some portion,
either positive or negative portion of this curve
to this mean curve.
And you can see over here how much of the variance
you can explain with that one curve.
So in that case, if we just had our mean curve
in our first shape,
we would explain about 50% of the variance.
By adding a second function, now we're explaining nearly 79%,
3rd function gets us up to 88%.
And so you can see how much of that variation we can explain
by adding more and more shapes.
And depending on the type of curve you have,
you might have only one function or you might have dozens of functions
depending on what that curve looks like.
So this is it looks like we can explain a lot of the variance here.
It takes us about nine functions to get up there.
But now we want to see how well those combinations
of all those shapes with the mean function or with the mean curve,
how those are represented.
How representative they are of our experimental data.
So to do that, we're going to go down to the score plot,
and I'm going to make this just a little bit smaller.
And we're going to look at the score plot and this FPC profiler.
So the FPC profiler
is a way to show the combination of all those different shapes.
So we really just want to pay attention to this top left part of the grid.
So this is our experiment based on the combination of the mean curve
with all those different shapes.
And right now, all of the FPCs, since they're set to zero,
we just get that mean curve.
But if I start adding that first FPC, if I make it more positive,
I'm adding that shape,
now I can see how my modeled shape is changing,
and if I go lower, I can see how it's changing.
So by adding and subtracting each of these different shapes,
I can recreate all of the different curves
or get close to all those different curves.
So this might take a little while to do manually,
but there's a nice little shortcut.
So what I like to do is go into this score plot,
and let's say I want to see experiment number six
so I can hover over six,
and then I'm going to pin that here, pull it over.
And so there are nine different functions, but we're only going to see two at a time.
And I can see that component one is 0.08
and component two is minus 0.03 .
So I can take this to 0.08 , and I can take this to minus 0.03 .
And I'm starting to reproduce this curve, but I would need to adjust all of them.
And so there's a shortcut to do that by just clicking on this.
So by clicking on six, all of the different FPC components
are set to the best representative model.
And we can look at these two shapes and see how close they are.
In this case, they look pretty good.
Maybe there's not as much definition, and this is not very straight,
but it looks pretty good.
And so we can go over to another curve like number seven,
we'll click on that one and see how it changes.
Now it's not looking quite as good, and we can go to eight.
And this is what I really like about this platform,
is it lets you explore the data.
So it's Functional D ata Explorer,
we're just seeing how well this model fits,
and we're doing it fairly visually.
And right here, if we're really interested in that understanding the bimodal nature
we're not getting that resolution with here.
So this is telling us maybe this isn't the best model.
Maybe there's a better one out there that we can look at.
So if we go up back to the top,
the linear model was selected initially because it had the minimum BIC,
but maybe we want to use a step function so I can click on the step function.
And now all those FPC curves have been recalculated.
And the first thing we notice is,
we're getting a lot more explanation of the variance here.
So we don't necessarily need all of these curves,
I can just take this slider.
Maybe we just want to look at six curves and explain 99.7% the variance.
And so it's simplifying the model a bit.
So now we can go down here and take another look
and spot check those curves.
So I can hover over six again, pin it here.
This is the curve that we'll be looking at.
And what I want to do, I'll just make this a little bit bigger.
And so when I click on six, okay so what do you think, Jerry?
What do you think this one is looking a little bit better?
-That's much better reproduction of your experimental day?
Yeah, I like that.
-Good. Yeah, I think this is looking better.
So we can go to seven,
and that one's looking a lot better as well.
And you don't need to select them all,
but it's good to check a few of them so we can go look at eight.
Yeah, so now we're getting a lot better resolution here
on the bimodal nature of it.
All right, I think this is telling us that this model is pretty good.
-Yeah, so what do we do now?
That's great that you can reproduce the experimental results,
but how do you get to the optimal?
-Yeah, I guess at this stage, it's still a little bit abstract.
So we've got all these different shapes that we're combining in different ways
to reproduce all of our curves, but we haven't done what we set out to do
which was relate those shapes to our DOE factors.
So that's what we're going to do next.
We're going to go back up to the model
and we're going to select functional DOE analysis.
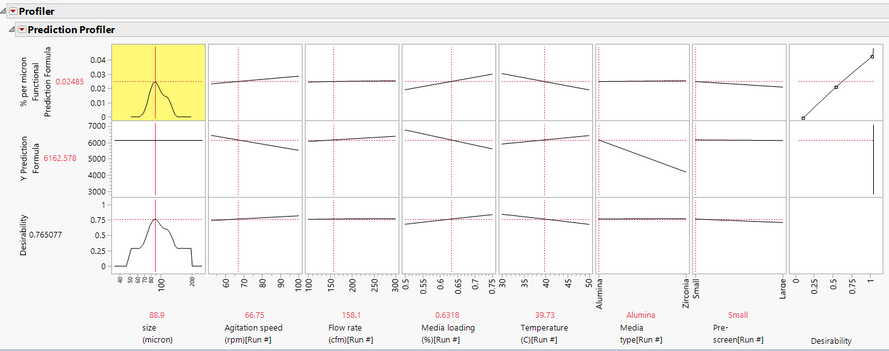
And when we do that, now we're getting a profiler
that might look a little bit more familiar if you're used to doing traditional DEO.
So once again, the response curve that we have is here.
And so we see our percent per micron on the Y
and the micron size or the particle size on the X.
But now instead of having those FPCs
in those shapes, now we have our DOE factors,
so we've got our agitation speed, our flow rate, media loading, et cetera.
Now I can move that agitation speed and I can see how it's relating to
or how it's influencing the curve.
And I can see by the slope of these lines
whether or not something is important or not.
So changing that one doesn't really change the shape.
So flow rate doesn't matter a whole lot, but temperature certainly
makes it more broad or makes it more narrow.
And so what we can do because we loaded that target curve,
just like in a standard deal, we can go to our red triangle
and we can go to maximize desirability.
So typically, this would look at a parameter
if you were doing a traditional DOE.
But in this case, it's going to try to find the settings
that match that target curve that we loaded earlier.
So when I click that, and there we go.
It looks like there are some settings here that get us a curve that's fairly narrow,
hitting the peak that we wanted and doesn't have any of those features
that we're trying to avoid.
-Very cool.
So are we done?
We've got the settings that we need, we just throw those
over the fence manufacturing and we're done.
-Well, that's one way to doe it, Jerry.
I don't know if folks in manufacturing that I worked with before
might not like that.
We probably want because these settings were set at,
the settings are not part of our design.
So this one is in the center,
this one's not at an edge or the center.
So we probably want to run some confirmation runs here,
maybe see some sensitivities and make sure that,,
we've got some robustness around these settings.
-Very cool.
Okay, all right.
-Let's get back and I think we can sum up.
-Yeah. So, Scott, that was great,
thank you for that presentation.
So in summary, what we've tried to do, is demonstrate how to perform a DOE
using these particle size density curves.
The curves themselves as the response rather than parameterizing the PSDs
with summary statistics like median, standard deviation, et cetera.
We were then able to optimize our factor input settings to the process
at least at the pilot scale to find an optimal curve shape that was very close
to our desired particle size distribution.
Along with the way we discovered how some of those parameters,
agitation speed and so forth affect the particle size distribution,
leading to multiple peaks or leading to broad peaks
or whatever that might be.
So we have an understanding about that, and we have a model.
So bringing this all back to our original scenario,
R& D took the results back to manufacturing,
where confirmation runs were attempted.
Scale- up perhaps wasn't completely successful.
That's typical of scale- ups, sometimes the pilot runs
don't map directly to manufacturing.
But we do have this model now that gives us an indication
of which of these knobs to turn to adjust if we do have a shoulder on that peak
or something like that.
So we were able to go back to manufacturing,
give them the assistance that they needed to get that in
so that they could increase their throughput and everyone was happy.
[crosstalk 00:26:18] That concludes our paper.
Scott, thanks for all the hard work.
-Yeah, well, it was great working with you on this, Jerry.
-Yeah, likewise.
Scott has been kind enough to save the modeling script in the data table,
which we're going to attach to this presentation.
If you've got any questions about the video
or any of the techniques that we did, please post your comments below the video,
there'll be a space for you to do that, we'd be happy to get back with you.
Thank you for joining us.
-Yap, thanks.